Working with linear solvers
Sparse LU decomposition (Gaussian elimination) is used by default to solve linear systems of equations in FEniCS programs. This is a very robust and simple method. It is the recommended method for systems with up to a few thousand unknowns and may hence be the method of choice for many 2D and smaller 3D problems. However, sparse LU decomposition becomes slow and one quickly runs out of memory for larger problems. For large problems, we instead need to use iterative methods which are faster and require much less memory. We will now look at how to take advantage of state-of-the-art iterative solution methods in FEniCS.
Controlling the solution process
Choosing a linear solver and preconditioner
Preconditioned Krylov solvers is a type of popular iterative methods that are easily accessible in FEniCS programs. The Poisson equation results in a symmetric, positive definite system matrix, for which the optimal Krylov solver is the Conjugate Gradient (CG) method. However, the CG method requires boundary conditions to be implemented in a symmetric way. This is not the case by default, so then a Krylov solver for non-symmetric system, such as GMRES, is a better choice. Incomplete LU factorization (ILU) is a popular and robust all-round preconditioner, so let us try the GMRES-ILU pair:
solve(a == L, u, bc)
solver_parameters={'linear_solver': 'gmres',
'preconditioner': 'ilu'})
# Alternative syntax
solve(a == L, u, bc,
solver_parameters=dict(linear_solver='gmres',
preconditioner='ilu'))
the section List of linear solver methods and preconditioners lists the most popular choices of Krylov solvers and preconditioners available in FEniCS.
Choosing a linear algebra backend
The actual GMRES and ILU implementations that are brought into action depend on the choice of linear algebra package. FEniCS interfaces several linear algebra packages, called linear algebra backends in FEniCS terminology. PETSc is the default choice if FEniCS is compiled with PETSc. If PETSc is not available, then FEniCS falls back to using the Eigen backend. The linear algebra backend in FEniCS can be set using the following command:
parameters['linear_algebra_backend'] = backendname
where backendname is a string. To see which linear algebra backends
are available, you can call the FEniCS function
list_linear_algebra_backends(). Similarly, one may check which
linear algebra backend is currently being used by the following
command:
print parameters['linear_algebra_backend']
# Alternative syntax for Python 3
print(parameters['linear_algebra_backend'])
Setting solver parameters
We will normally want to control the tolerance in the stopping criterion and the maximum number of iterations when running an iterative method. Such parameters can be controlled at both a global and a local level. We will start by looking at how to set global parameters. For more advanced programs, one may want to use a number of different linear solvers and set different tolerances and other parameters. Then it becomes important to control the parameters at a local level. We will return to this issue in the section Linear variational problem and solver objects.
Changing a parameter in the global FEniCS parameter database affects
all linear solvers (created after the parameter has been set).
The global FEniCS parameter database is simply called parameters and
it behaves as a nested dictionary. Write
info(parameters, verbose=True)
to list all parameters and their default values in the database.
The nesting of parameter sets is indicated through indentation in the
output from info.
According to this output, the relevant parameter set is
named 'krylov_solver', and the parameters are set like this:
prm = parameters['krylov_solver'] # short form
prm['absolute_tolerance'] = 1E-10
prm['relative_tolerance'] = 1E-6
prm['maximum_iterations'] = 1000
Stopping criteria for Krylov solvers usually involve the norm of the residual, which must be smaller than the absolute tolerance parameter or smaller than the relative tolerance parameter times the initial residual.
We remark that default values for the global parameter database can be defined in an XML file. To generate such a file from the current set of parameters in a program, run
File('fenics_parameters.xml') << parameters
If a fenics_parameters.xml file is found in the directory where a
FEniCS program is run, this file is read and used to initialize the
parameters object. Otherwise, the file
.config/fenics/fenics_parameters.xml in the user's home directory is
read, if it exists. Another alternative is to load the XML file (with any
name) manually in the program:
File('fenics_parameters.xml') >> parameters
The XML file can also be in gzip'ed form with the extension .xml.gz.
An extended solver function
We may extend the previous solver function from
ft13_poisson_solver.py in the section A more general solver function
such that it also offers the GMRES+ILU
preconditioned Krylov solver:
def solver(
f, u_b, Nx, Ny,
degree=1, # Polynomial degree of function space
linear_solver='Krylov', # Linear solver method (alt: 'direct')
abs_tol=1E-5, # Absolute tolerance in Krylov solver
rel_tol=1E-3, # Relative tolerance in Krylov solver
max_iter=1000, # Max iterations in Krylov solver
log_level=PROGRESS, # Amount of solver output
print_parameters=False, # Print solver parameters to screen?
):
...
# Set up variational problem: a, L, declare u, etc.
if linear_solver == 'Krylov':
prm = parameters['krylov_solver']
prm['absolute_tolerance'] = abs_tol
prm['relative_tolerance'] = rel_tol
prm['maximum_iterations'] = max_iter
print(parameters['linear_algebra_backend'])
set_log_level(log_level)
if print_parameters:
info(parameters, True)
solver_parameters = {'linear_solver': 'gmres',
'preconditioner': 'ilu'}
else:
solver_parameters = {'linear_solver': 'lu'}
solve(a == L, u, bc, solver_parameters=solver_parameters)
return u
This new solver function, found in the file
ft15_poisson_iterative.py, replaces the one in ft13_poisson_solver.py:
it has all the functionality of the previous solver function,
but can also solve the linear system with
iterative methods and report the progress of such solvers.
A remark regarding unit tests
Regarding verification of the new solver function in terms of unit
tests, it turns out that unit testing for a problem where the
approximation error vanishes gets more complicated when we use
iterative methods. The problem is to keep the error due to iterative
solution smaller than the tolerance used in the verification
tests. First of all, this means that the tolerances used in the Krylov
solvers must be smaller than the tolerance used in the assert test,
but this is no guarantee to keep the linear solver error this small.
For linear elements and small meshes, a tolerance of \( 10^{-11} \) works
well in the case of Krylov solvers too (using a tolerance \( 10^{-12} \)
in those solvers). However, as soon as we switch to \( \mathsf{P}_2 \)
elements, it is hard to force the linear solver error below
\( 10^{-6} \). Consequently, tolerances in tests depend on the numerical
method being used. The interested reader is referred to the
test_solver function in ft15_poisson_iterative.py for details:
this function tests the numerical solution for direct and iterative
linear solvers, for different meshes, and different degrees of the
polynomials in the finite element basis functions.
List of linear solver methods and preconditioners
Which linear solvers and preconditioners that are available in FEniCS depends on how FEniCS has been configured and which linear algebra backend is currently active. The following table shows an example of which linear solvers that can be available through FEniCS when the PETSc backend is active:
| Name | Method |
'bicgstab' | Biconjugate gradient stabilized method |
'cg' | Conjugate gradient method |
'gmres' | Generalized minimal residual method |
'minres' | Minimal residual method |
'petsc' | PETSc built in LU solver |
'richardson' | Richardson method |
'superlu_dist' | Parallel SuperLU |
'tfqmr' | Transpose-free quasi-minimal residual method |
'umfpack' | UMFPACK |
The set of available preconditioners also depends on configuration and linear algebra backend. The following table shows an example of which preconditioners may be available:
| Name | Method |
'icc' | Incomplete Cholesky factorization |
'ilu' | Incomplete LU factorization |
'petsc_amg' | PETSc algebraic multigrid |
'sor' | Successive over-relaxation |
An up-to-date list of the available solvers and preconditioners for your FEniCS installation can be produced by
list_linear_solver_methods()
list_krylov_solver_preconditioners()
Linear variational problem and solver objects
The FEniCS interface allows different ways to access the core
functionality, ranging from very high-level to low-level access. So
far, we have mostly used the high-level call solve(a == L, u, bc) to
solve a variational problem a == L with a certain boundary condition
bc. However, sometimes you may need more fine-grained control over
the solution process. In particular, the call to solve will create
certain objects that are thrown away after the solution has been
computed, and it may be practical or efficient to reuse those
objects.
In this section, we will look at an alternative interface to solving
linear variational problems in FEniCS, which may be preferable in
many situations compared to the high-level solve function interface.
This interface uses the two classes LinearVariationalProblem and
LinearVariationalSolver. Using this interface, the equivalent of
solve(a == L, u, bc) looks as follows:
u = Function(V)
problem = LinearVariationalProblem(a, L, u, bc)
solver = LinearVariationalSolver(problem)
solver.solve()
Many FEniCS objects have an attribute parameters corresponding to
a parameter set in the global parameters database,
but local to the object. Here, solver.parameters play that
role. Setting the CG method with ILU preconditioning as the solution
method and specifying solver-specific parameters can be done
like this:
solver.parameters['linear_solver'] = 'gmres'
solver.parameters['preconditioner'] = 'ilu'
prm = solver.parameters['krylov_solver'] # short form
prm['absolute_tolerance'] = 1E-7
prm['relative_tolerance'] = 1E-4
prm['maximum_iterations'] = 1000
Settings in the global parameters database are
propagated to parameter sets in individual objects, with the
possibility of being overwritten as above. Note that global parameter
values can only affect local parameter values if set before the time
of creation of the local object. Thus, changing the value of the
tolerance in the global parameter database will not affect the
parameters for already created solvers.
The linear variational problem and solver objects as outlined above
are incorporated in an alternative solver function, named
solver_objects, in
ft15_poisson_iterative.py. Otherwise, this function is similar to the
previously shown solver function.
Explicit assembly and solve
As we saw already in the section The Navier--Stokes equations, linear variational
problems can be assembled explicitly in FEniCS into matrices and
vectors using the assemble function. This allows even more
fine-grained control of the solution process compared to using the
high-level solve function or using the classes
LinearVariationalProblem and
LinearVariationalSolver. We will now look more closely into how to
use the assemble function and how to combine this with low-level
calls for solving the assembled linear systems.
Given a variational problem \( a(u,v)=L(v) \), the discrete solution \( u \) is computed by inserting \( u=\sum_{j=1}^N U_j \phi_j \) into \( a(u,v) \) and demanding \( a(u,v)=L(v) \) to be fulfilled for \( N \) test functions \( \hat\phi_1,\ldots,\hat\phi_N \). This implies $$ \begin{equation*} \sum_{j=1}^N a(\phi_j,\hat\phi_i) U_j = L(\hat\phi_i),\quad i=1,\ldots,N, \end{equation*} $$ which is nothing but a linear system, $$ \begin{equation*} AU = b, \end{equation*} $$ where the entries of \( A \) and \( b \) are given by $$ \begin{align*} A_{ij} &= a(\phi_j, \hat{\phi}_i), \\ b_i &= L(\hat\phi_i)\tp \end{align*} $$
The examples so far have specified the left- and right-hand sides of
the variational formulation and then asked FEniCS to assemble the
linear system and solve it. An alternative is to explicitly call
functions for assembling the coefficient matrix \( A \) and the right-hand
side vector \( b \), and then solve the linear system \( AU=b \) for
the vector \( U \). Instead of solve(a == L, U, b) we now write
A = assemble(a)
b = assemble(L)
bc.apply(A, b)
u = Function(V)
U = u.vector()
solve(A, U, b)
The variables a and L are the same as before; that is, a refers
to the bilinear form involving a TrialFunction object u
and a TestFunction object v, and L involves the same TestFunction
object v. From a and L, the assemble function can compute
\( A \) and \( b \).
Creating the linear system explicitly in a program can have some advantages in more advanced problem settings. For example, \( A \) may be constant throughout a time-dependent simulation, so we can avoid recalculating \( A \) at every time level and save a significant amount of simulation time.
The matrix \( A \) and vector \( b \) are first assembled without
incorporating essential (Dirichlet) boundary conditions. Thereafter,
the call bc.apply(A, b) performs the necessary modifications of the
linear system such that u is guaranteed to equal the prescribed
boundary values. When we have multiple Dirichlet conditions stored in
a list bcs, we must apply each condition in bcs to the system:
for bc in bcs:
bc.apply(A, b)
# Alternative syntax using list comprehension
[bc.apply(A, b) for bc in bcs]
Alternatively, we can use the function assemble_system, which takes
the boundary conditions into account during the assembly of the matrix
and vector:
A, b = assemble_system(a, L, bcs)
The assemble_system function is preferable to the combination of
assemble and bc.apply when the linear system is symmetric, since
assemble_system will incorporate the boundary conditions in a
symmetric way. Even if the matrix A that comes out of the call to
assemble is symmetric (for a symmetric bilinear form a), the call to bc.apply
will break the symmetry.
Once the linear system has been assembled, we need to compute the
solution \( U=A^{-1}b \) and store the solution \( U \) in the vector
U = u.vector(). In the same way as linear variational problems can be
programmed using different interfaces in FEniCS—the high-level
solve function, the class LinearVariationalSolve, and the
low-level assemble function—linear systems can also be programmed
using different interfaces in FEniCS. The high-level interface to
solving a linear system in FEniCS is also named solve:
solve(A, U, b)
By default, solve(A, U, b) uses sparse LU decomposition to compute
the solution. Specification of an iterative solver and preconditioner
can be made through two optional arguments:
solve(A, U, b, 'cg', 'ilu')
Appropriate names of solvers and preconditioners are found in the section List of linear solver methods and preconditioners.
This high-level interface is useful for many applications, but
sometimes more fine-grained control is needed. One can then create one
or more KrylovSolver objects that are then used to solve linear
systems. Each different solver object can have its own set of
parameters and selection of iterative method and preconditioner. Here
is an example:
solver = KrylovSolver('cg', 'ilu')
prm = solver.parameters
prm['absolute_tolerance'] = 1E-7
prm['relative_tolerance'] = 1E-4
prm['maximum_iterations'] = 1000
u = Function(V)
U = u.vector()
solver.solve(A, U, b)
The function solver_linalg in the program file
ft15_poisson_iterative.py implements a solver function where the user
can choose between different types of assembly. The function
demo_linalg runs a test problem on a sequence of meshes and
solves the problem with symmetric and non-symmetric modification of
the system matrix. One can monitor the number of Krylov method
iterations and realize that with a symmetric coefficient matrix, the
Conjugate Gradient method requires slightly fewer iterations than
GMRES in the non-symmetric case. Taking into account that the
Conjugate Gradient method has less work per iteration, there is some
efficiency to be gained by using assemble_system for this problem.
The choice of start vector for the iterations in a linear solver is
often important. By default, the values of u and thus the vector U
= u.vector() will be initialized to zero. If we instead wanted to
initialize U with random numbers in the interval \( [-100,100] \) this
can be done as follows:
n = u.vector().array().size
U = u.vector()
U[:] = numpy.random.uniform(-100, 100, n)
solver.parameters['nonzero_initial_guess'] = True
solver.solve(A, U, b)
Note that we must both turn off the default behavior of setting the start
vector ("initial guess") to zero, and also set the values of the
vector U to nonzero values.
Using a nonzero initial guess can be particularly important for
time-dependent problems or when solving a linear system as part of a
nonlinear iteration, since then the previous solution vector U will
often be a good initial guess for the solution in the next time step
or iteration. In this case, the values in the vector U will
naturally be initialized with the previous solution vector (if we just
used it to solve a linear system), so the only extra step necessary is
to set the parameter nonzero_initial_guess to True.
Examining matrix and vector values
When calling A = assemble(a) and b = assemble(L), the object A
will be of type Matrix, while b and u.vector() are of type
Vector. To examine the values, we may convert the matrix and vector
data to numpy arrays by calling the array() method as shown
before. For example, if you wonder how essential boundary conditions are
incorporated into linear systems, you can print out A and b
before and after the bc.apply(A, b) call:
A = assemble(a)
b = assemble(L)
if mesh.num_cells() < 16: # print for small meshes only
print(A.array())
print(b.array())
bc.apply(A, b)
if mesh.num_cells() < 16:
print(A.array())
print(b.array())
With access to the elements in A through a numpy array, we can easily
perform computations on this matrix, such as computing the eigenvalues
(using the eig function in numpy.linalg). We can alternatively dump
A.array() and b.array() to file in MATLAB format and invoke
MATLAB or Octave to analyze the linear system.
Dumping the arrays to MATLAB format is done by
import scipy.io
scipy.io.savemat('Ab.mat', {'A': A.array(), 'b': b.array()})
Writing load Ab.mat in MATLAB or Octave will then make
the array variables A and b available for computations.
Matrix processing in Python or MATLAB/Octave is only feasible for
small PDE problems since the numpy arrays or matrices in MATLAB file
format are dense matrices. FEniCS also has an interface to the
eigensolver package SLEPc, which is the preferred tool for computing
the eigenvalues of large, sparse matrices of the type encountered in
PDE problems (see demo/documented/eigenvalue in the FEniCS source
code tree for a demo).
Examining the degrees of freedom
We have seen before how to grab the degrees of freedom array from a
finite element function u:
nodal_values = u.vector().array()
For a finite element function from a standard continuous piecewise linear function space (\( \mathsf{P}_1 \) Lagrange elements), these values will be the same as the values we get by the following statement:
vertex_values = u.compute_vertex_values(mesh)
Both nodal_values and vertex_values will be numpy arrays and they will
be of the same length and contain the same values, but with possibly
different ordering. The array vertex_values will have the same
ordering as the vertices of the mesh, while nodal_values will be ordered in
a way that (nearly) minimizes the bandwidth of the system matrix and
thus improves the efficiency of linear solvers.
A fundamental question is: what are the
coordinates of the vertex whose value is nodal_values[i]? To answer this
question, we need to understand how to get our hands on the
coordinates, and in particular, the numbering of degrees of freedom
and the numbering of vertices in the mesh.
The function mesh.coordinates() returns the coordinates of the
vertices as a numpy array with shape \( (M,d \)), \( M \) being the number
of vertices in the mesh and \( d \) being the number of space dimensions:
>>> from fenics import *
>>>
>>> mesh = UnitSquareMesh(2, 2)
>>> coordinates = mesh.coordinates()
>>> coordinates
array([[ 0. , 0. ],
[ 0.5, 0. ],
[ 1. , 0. ],
[ 0. , 0.5],
[ 0.5, 0.5],
[ 1. , 0.5],
[ 0. , 1. ],
[ 0.5, 1. ],
[ 1. , 1. ]])
We see from this output that for this particular mesh, the vertices are first numbered along \( y=0 \) with increasing \( x \) coordinate, then along \( y=0.5 \), and so on.
Next we compute a function u on this mesh. Let's take \( u=x+y \):
>>> V = FunctionSpace(mesh, 'P', 1)
>>> u = interpolate(Expression('x[0] + x[1]'), V)
>>> plot(u, interactive=True)
>>> nodal_values = u.vector().array()
>>> nodal_values
array([ 1. , 0.5, 1.5, 0. , 1. , 2. , 0.5, 1.5, 1. ])
We observe that nodal_values[0] is not the value of \( x+y \) at
vertex number 0, since this vertex has coordinates \( x=y=0 \). The
numbering of the nodal values (degrees of freedom) \( U_1,\ldots,U_{N} \)
is obviously not the same as the numbering of the vertices.
Note that we may examine the vertex numbering in the plot window. We
type w to turn on wireframe instead of a fully colored surface, m
to show the mesh, and then v to show the numbering of the vertices.
Let's instead examine the values we get by calling
u.compute_vertex_values():
>>> vertex_values = u.compute_vertex_values()
>>> for i, x in enumerate(coordinates):
... print('vertex %d: vertex_values[%d] = %g\tu(%s) = %g' %
... (i, i, vertex_values[i], x, u(x)))
vertex 0: vertex_values[0] = 0 u([ 0. 0.]) = 8.46545e-16
vertex 1: vertex_values[1] = 0.5 u([ 0.5 0. ]) = 0.5
vertex 2: vertex_values[2] = 1 u([ 1. 0.]) = 1
vertex 3: vertex_values[3] = 0.5 u([ 0. 0.5]) = 0.5
vertex 4: vertex_values[4] = 1 u([ 0.5 0.5]) = 1
vertex 5: vertex_values[5] = 1.5 u([ 1. 0.5]) = 1.5
vertex 6: vertex_values[6] = 1 u([ 0. 1.]) = 1
vertex 7: vertex_values[7] = 1.5 u([ 0.5 1. ]) = 1.5
vertex 8: vertex_values[8] = 2 u([ 1. 1.]) = 2
We can ask FEniCS to give us the mapping from vertices to degrees of freedom for a certain function space \( V \):
v2d = vertex_to_dof_map(V)
Now, nodal_values[v2d[i]] will give us the value of the degree of freedom in
u corresponding to vertex i (v2d[i]). In particular, nodal_values[v2d]
is an array with all the elements in the same (vertex numbered) order
as coordinates. The inverse map, from degrees of freedom number to
vertex number is given by dof_to_vertex_map(V). This means that
we may call
coordinates[dof_to_vertex_map(V)] to get an array of all the
coordinates in the same order as the degrees of freedom. Note that
these mappings are only available in FEniCS for \( \mathsf{P}_1 \) elements.
For Lagrange elements of degree larger than 1, there are degrees of
freedom (nodes) that do not correspond to vertices. For these
elements, we may get the vertex values by
calling u.compute_vertex_values(mesh), and we can get the degrees of
freedom by calling u.vector().array(). To get the coordinates
associated with all degrees of freedom, we need to iterate over the
elements of the mesh and ask FEniCS to return the coordinates and dofs
associated with each cell. This information is stored in the
FiniteElement and DofMap object of a FunctionSpace. The
following code illustrates how to iterate over all elements of a mesh and
print the degrees of freedom and coordinates associated with the
element.
element = V.element()
dofmap = V.dofmap()
for cell in cells(mesh):
print(element.tabulate_dof_coordinates(cell))
print(dofmap.cell_dofs(cell.index()))
Function object u, we can evaluate its values in various
ways:
-
u(x)for an arbitrary pointx -
u.vector().array()[i]for degree of freedom numberi -
u.compute_vertex_values()[i]at vertex numberi
To demonstrate the use of point evaluation of Function objects, we
write out the computed u at the center point of the domain and
compare it with the exact solution:
center = (0.5, 0.5)
error = u_D(center) - u(center)
print('Error at %s: %g' % (center, error))
For a \( 2\times(3\times 3) \) mesh, the output from the previous snippet becomes
Error at (0.5, 0.5): -0.0833333
The discrepancy is due to the fact that the center point is not a node
in this particular mesh, but a point in the interior of a cell, and
u varies linearly over the cell while u_D is a quadratic
function. When the center point is a node, as in a \( 2\times(2\times
2) \) or \( 2\times(4\times 4) \) mesh, the error is of the order
\( 10^{-15} \).
We have seen how to extract the nodal values in a numpy array.
If desired, we can adjust the nodal values too. Say we want to
normalize the solution such that \( \max_j |U_j| = 1 \). Then we
must divide all \( U_j \) values
by \( \max_j |U_j| \). The following function performs the task:
def normalize_solution(u):
"""Normalize solution by dividing by max(|u|)."""
nodal_values = u.vector().array()
u_max = np.abs(nodal_values).max()
nodal_values /= u_max
u.vector()[:] = nodal_values
#u.vector().set_local(dofs) # alternative
The /= operator implies an
in-place modification of the object on the left-hand side: all
elements of the array nodal_values are divided by the value u_max.
Alternatively, we could do nodal_values = nodal_values / u_max, which
implies creating a new array on the right-hand side and assigning this
array to the name nodal_values.
u.vector().array() returns a copy of the data in
u.vector(). One must therefore never perform assignments like
u.vector.array()[:] = ..., but instead extract the numpy array
(i.e., a copy), manipulate it, and insert it back with u.vector()[:]
= or use u.set_local(...).
All the code in this can be found in the file
ft15_poisson_iterative.py (Poission solver with use of iterative
methods).
Postprocessing computations
As the final theme in this chapter, we will look at how to postprocess computations; that is, how to compute various derived quantities from the computed solution of a PDE. The solution \( u \) itself may be of interest for visualizing general features of the solution, but sometimes one is interested in computing the solution of a PDE to compute a specific quantity that derives from the solution, such as, e.g., the flux, a point-value, or some average of the solution.
A variable-coefficient Poisson problem
As a test problem, we will extend the Poisson problem from the chapter Fundamentals: Solving the Poisson equation with a variable coefficient \( \kappa(x,y) \) in the Laplace operator: $$ \begin{alignat}{2} - \nabla\cdot \left\lbrack \kappa(x,y)\nabla u(x,y)\right\rbrack &= f(x,y) \quad &\mbox{in } \Omega, \tag{5.3} \\ u(x,y) &= \ub(x,y) \quad &\mbox{on}\ \partial\Omega\tp \end{alignat} $$ Let us continue to use our favorite solution \( u(x,y)=1+x^2+2y^2 \) and then prescribe \( \kappa(x,y)=x+y \). It follows that \( \ub(x,y) = 1 + x^2 + 2y^2 \) and \( f(x,y)=-8x-10y \).
We shall quickly demonstrate that this simple extension of our model problem only requires an equally simple extension of the FEniCS program. The following simple changes must be made to the previously shown codes:
- the
solverfunction must takek(\( \kappa \)) as an argument, - a new
Expression kmust be defined for the variable coefficient, - the right-hand side
fmust be anExpressionsince it is no longer a constant, - the formula for \( a(u,v) \) in the variational problem must be updated.
a = dot(grad(u), grad(v))*dx
by
a = k*dot(grad(u), grad(v))*dx
Moreover,
the definitions of k and f in the test problem read
k = Expression('x[0] + x[1]')
f = Expression('-8*x[0] - 10*x[1]')
No additional modifications are necessary. The file
ft11_poisson_bcs.py (Poisson problem, variable coefficients)
is a copy of ft15_poisson_iterative.py with the mentioned changes
incorporated. Observe that \( \kappa=1 \) recovers the original problem in
ft15_poisson_iterative.py.
You can execute the file and confirm that it recovers the exact solution \( u \) at the nodes.
Flux computations
It is often of interest to compute the flux \( Q = -\kappa\nabla u \). Since \( u = \sum_{j=1}^N U_j \phi_j \), it follows that $$ \begin{equation*} Q = -\kappa\sum_{j=1}^N U_j \nabla \phi_j\tp \end{equation*} $$
However, the gradient of a piecewise continuous finite element scalar field is a discontinuous vector field since the basis functions \( \{\phi_j\} \) have discontinuous derivatives at the boundaries of the cells. For example, using Lagrange elements of degree 1, \( u \) is linear over each cell, and the gradient becomes a piecewise constant vector field. On the contrary, the exact gradient is continuous. For visualization and data analysis purposes, we often want the computed gradient to be a continuous vector field. Typically, we want each component of \( \nabla u \) to be represented in the same way as \( u \) itself. To this end, we can project the components of \( \nabla u \) onto the same function space as we used for \( u \). This means that we solve \( w = \nabla u \) approximately by a finite element method, using the same elements for the components of \( w \) as we used for \( u \). This process is known as projection.
Projection is a common operation in finite element analysis and FEniCS
has a function for easily performing the projection:
project(expression, W), which returns the projection of some
expression into the space W.
In our case, the flux \( Q = -\kappa\nabla u \)
is vector-valued and we need to pick W as the vector-valued function
space of the same degree as the space V where u resides:
V = u.function_space()
mesh = V.mesh()
degree = V.ufl_element().degree()
W = VectorFunctionSpace(mesh(), 'P', degree)
grad_u = project(grad(u), W)
flux_u = project(-k*grad(u), W)
The applications of projection are many, including turning discontinuous gradient fields into continuous ones, comparing higher- and lower-order function approximations, and transforming a higher-order finite element solution down to a piecewise linear field, which is required by many visualization packages.
Plotting the flux vector field is naturally as easy as plotting anything else:
plot(flux_u, title='flux field')
flux_x, flux_y = flux_u.split(deepcopy=True) # extract components
plot(flux_x, title='x-component of flux (-kappa*grad(u))')
plot(flux_y, title='y-component of flux (-kappa*grad(u))')
The deepcopy=True argument signifies a deep copy, which is
a general term in computer science implying that a copy of the data is
returned. (The opposite, deepcopy=False,
means a shallow copy, where
the returned objects are just pointers to the original data.)
For data analysis of the nodal values of the flux field, we can
grab the underlying numpy arrays (which demands a deepcopy=True
in the split of flux):
flux_x_nodal_values = flux_x.vector().dofs()
flux_y_nodal_values = flux_y.vector().dofs()
The degrees of freedom of the flux_u vector field can also be
reached by
flux_u_nodal_values = flux_u.vector().array()
However, this is a flat numpy array containing the degrees of
freedom for both the \( x \) and \( y \) components of the flux and the
ordering of the components may be mixed up by FEniCS in order to
improve computational efficiency.
The function demo_test_flux in the program
ft11_poisson_bcs.py demonstrates the computations described
above.
project to project a finite element
function, it can be instructive to look at how to formulate the
projection mathematically and implement its steps manually in FEniCS.
Let's say we have an expression \( g = g(u) \) that we want to project into some space \( W \). The mathematical formulation of the (\( L^2 \)) projection \( w = P_W g \) into \( W \) is the variational problem $$ \begin{equation} \int_{\Omega} w v \dx = \int_{\Omega} g v \dx \tag{5.5} \end{equation} $$ for all test functions \( v\in W \). In other words, we have a standard variational problem \( a(w, v) = L(v) \) where now $$ \begin{align} a(w, v) &= \int_\Omega w v \dx, \tag{5.6}\\ L(v) &= \int_\Omega g v \dx\tp \tag{5.7} \end{align} $$ Note that when the functions in \( W \) are vector-valued, as is the case when we project the gradient \( g(u) = \nabla u \), we must replace the products above by \( w\cdot v \) and \( g\cdot v \).
The variational problem is easy to define in FEniCS.
w = TrialFunction(W)
v = TestFunction(W)
a = w*v*dx # or dot(w, v)*dx when w is vector-valued
L = g*v*dx # or dot(g, v)*dx when g is vector-valued
w = Function(W)
solve(a == L, w)
The boundary condition argument to solve is dropped since there are
no essential boundary conditions in this problem.
Computing functionals
After the solution \( u \) of a PDE is computed, we occasionally want to compute functionals of \( u \), for example, $$ \begin{equation} {1\over2}||\nabla u||^2 = {1\over2}\int_\Omega \nabla u\cdot \nabla u \dx, \tag{5.8} \end{equation} $$ which often reflects some energy quantity. Another frequently occurring functional is the error $$ \begin{equation} ||\uex-u|| = \left(\int_\Omega (\uex-u)^2 \dx\right)^{1/2}, \tag{5.9} \end{equation} $$ where \( \uex \) is the exact solution. The error is of particular interest when studying convergence properties of finite element methods. Other times, we may instead be interested in computing the flux out through a part \( \Gamma \) of the boundary \( \partial\Omega \), $$ \begin{equation} F = -\int_\Gamma \kappa\nabla u\cdot n \ds, \tag{5.10} \end{equation} $$ where \( n \) is an outward unit normal at \( \Gamma \) and \( \kappa \) is a coefficient (see the problem in the section Postprocessing computations for a specific example).
All these functionals are easy to compute with FEniCS, as we shall see in the examples below.
Energy functional
The integrand of the energy functional (5.8) is described in the UFL language in the same manner as we describe weak forms:
energy = 0.5*dot(grad(u), grad(u))*dx
E = assemble(energy)
The functional energy is evaluated by calling the assemble
function that we have previously used to assemble matrices and
vectors. FEniCS will recognize that the form has ''rank 0'' (since it
contains no trial and test functions) and return the result as a
scalar value.
Error functional
Computing the functional (5.9) can be done as follows: by
error = (u_e - u)**2*dx
E = sqrt(abs(assemble(error)))
The exact solution \( \uex \) is here in a Function or Expression
object u_e, while u is the finite element approximation (and
thus a Function).
Sometimes, for very small error values, the result of
assemble(error) can be a (very small) negative number, so we have
used abs in the expression for E above to ensure a positive value
for the sqrt function.
As will be explained and demonstrated in the section Computing convergence rates, the integration of (u_e - u)**2*dx
can result in too optimistic convergence rates unless one is careful
how u_e is transferred onto a mesh. The general recommendation
for reliable error computation is to use the errornorm function (see
help(errornorm) and the section Computing convergence rates for
more information):
E = errornorm(u_e, u)
Flux Functional
To compute flux integrals like \( F = -\int_\Gamma \kappa\nabla u\cdot n \ds \), we need to define the \( n \) vector, referred to as facet normal in FEniCS. If the surface domain \( \Gamma \) in the flux integral is the complete boundary, we can perform the flux computation by
n = FacetNormal(mesh)
flux = -k*dot(grad(u), n)*ds
total_flux = assemble(flux)
Although grad(u) and nabla_grad(u) are interchangeable in the above
expression when u is a scalar function, we have chosen to write
grad(u) because this is the right expression if we generalize the
underlying equation to a vector Laplace/Poisson PDE. With nabla_grad(u) we
must in that case write dot(n, nabla_grad(u)).
It is possible to restrict the integration to a part of the boundary
by using a mesh function to mark the relevant part, as explained in
the section Setting multiple Dirichlet, Neumann, and Robin conditions. Assuming that the part corresponds
to subdomain number i, the relevant syntax for the variational
formulation of the flux is -k*dot(grad(u), n)*ds(i).
Computing convergence rates
A central question for any numerical method is its convergence rate: how fast does the error approach zero when the resolution is increased? For finite element methods, this typically corresponds to proving, theoretically or empirically, that the error \( e = \uex - u \) is bounded by the mesh size \( h \) to some power \( r \); that is, \( \|e\| \leq C h^r \) for some constant \( C \). The number \( r \) is called the convergence rate of the method. Note that different norms, like the \( L^2 \)-norm \( \|e\| \) or \( H^1_0 \)-norm \( \|\nabla e\| \) typically have different convergence rates.
To illustrate how to compute errors and convergence rates in FEniCS,
we have included the function convergence_rate in
ft11_poisson_bcs.py. This is a tool that is very handy
when verifying finite element codes and will therefore be explained in
detail here.
Computing error norms
As we have already seen, the \( L^2 \)-norm of the error \( \u_e - u \) can be implemented in FEniCS by
error = (u_e - u)**2*dx
E = sqrt(abs(assemble(error)))
As above, we have used abs in the expression for E above to ensure
a positive value for the sqrt function.
It is important to understand how FEniCS computes the error from the
above code, since we may otherwise run into subtle issues when using
the value for computing convergence rates. The first subtle issue is
that if u_e is not already a finite element function (an object created
using Function(V)), which is the case if u_e is defined as an
Expression, FEniCS must interpolate u_e into some local finite
element space on each element of the mesh. The degree used for the
interpolation is determined by the mandatory keyword argument to the
Expression class, for example:
u_e = Expression('sin(x[0])', degree=1)
This means that the error computed will not be equal to the actual error \( \|\uex - u\| \) but rather the difference between the finite element solution \( u \) and the piecewise linear interpolant of \( \uex \). This may yield a too optimistic (too small) value for the error. A better value may be achieved by interpolating the exact solution into a higher-order function space, which can be done by simply increasing the degree:
u_e = Expression('sin(x[0])', degree=3)
The second subtle issue is that when FEniCS evaluates the expression
(u_e - u)**2, this will be expanded into u_e**2 + u**2 -
2*u_e*u. If the error is small (and the solution itself is of
moderate size), this calculation will correspond to the subtraction of
two positive numbers (u_e**2 + u**2 \( \sim 1 \) and 2*u_e*u \( \sim
1 \)) yielding a small number. Such a computation is very prone to
round-off errors, which may again lead to an unreliable value for the
error. To make this situation worse, FEniCS may expand this
computation into a large number of terms, in particular for higher
order elements, making the computation very unstable.
To help with these issues, FEniCS provides the built-in function
errornorm which computes the error norm in a more intelligent
way. First, both u_e and u are interpolated into a higher-order
function space. Then, the degrees of freedom of u_e and u are
subtracted to produce a new function in the higher-order function
space. Finally, FEniCS integrates the square of the difference
function to get the value of the error norm. Using the errornorm
function is simple:
E = errornorm(u_e, u, normtype='L2')
It is illustrative to look at a short implementation of errornorm:
def errornorm(u_e, u):
V = u.function_space()
mesh = V.mesh()
degree = V.ufl_element().degree()
W = FunctionSpace(mesh, 'P', degree + 3)
u_e_W = interpolate(u_e, W)
u_W = interpolate(u, W)
e_W = Function(W)
e_W.vector()[:] = u_e_W.vector().array() - u_W.vector().array()
error = e_W**2*dx
return sqrt(abs(assemble(error)))
Sometimes it is of interest to compute the error of the
gradient field: \( ||\nabla (\uex-u)|| \),
often referred to as the \( H^1_0 \) or \( H^1 \) seminorm of the error.
This can either be expressed as above, replacing the expression for
error by error = dot(grad(e_W), grad(e_W))*dx, or by calling
errornorm in FEniCS:
E = errornorm(u_e, u, norm_type='H10')
Type help(errornorm) in Python for more information about available
norm types.
The function compute_errors in ft11_poisson_bcs.py
illustrates the computation of various error norms in FEniCS.
Computing convergence rates
Let's examine how to compute convergence rates in FEniCS.
The solver function in ft11_poisson_bcs.py allows us to
easily compute solutions
for finer and finer meshes and enables us to
study the convergence rate. Define the element size \( h=1/n \), where \( n \)
is the number of cell divisions in the \( x \) and \( y \) directions (n=Nx=Ny in
the code). We perform experiments with \( h_0>h_1>h_2>\cdots \) and compute
the corresponding errors \( E_0, E_1, E_2 \) and so forth. Assuming
\( E_i=Ch_i^r \) for unknown constants \( C \) and \( r \), we can compare two
consecutive experiments, \( E_{i-1}=Ch_{i-1}^r \) and \( E_i=Ch_i^r \), and
solve for \( r \):
$$
\begin{equation*}
r = {\ln(E_i/E_{i-1})\over\ln (h_i/h_{i-1})}\tp
\end{equation*}
$$
The \( r \) values should approach the expected convergence rate
(typically the polynomial degree + 1 for the \( L^2 \)-error) as \( i \)
increases.
The procedure above can easily be turned into Python code. Here
we run through a list of element degrees (\( \mathsf{P}_1 \), \( \mathsf{P}_2 \), \( \mathsf{P}_3 \), and \( \mathsf{P}_4 \)),
perform experiments over a series of refined meshes, and for
each experiment report the six error types as returned by compute_errors:
def convergence_rate(u_exact, f, u_D, kappa):
"""
Compute convergence rates for various error norms for a
sequence of meshes and elements.
"""
h = {} # discretization parameter: h[degree][level]
E = {} # error measure(s): E[degree][level][error_type]
degrees = 1, 2, 3, 4
num_levels = 5
# Iterate over degrees and mesh refinement levels
for degree in degrees:
n = 4 # coarsest mesh division
h[degree] = []
E[degree] = []
for i in range(num_levels):
n *= 2
h[degree].append(1.0 / n)
u = solver(kappa, f, u_D, n, n, degree,
linear_solver='direct')
errors = compute_errors(u_exact, u)
E[degree].append(errors)
print('2 x (%d x %d) P%d mesh, %d unknowns, E1=%g' %
(n, n, degree, u.function_space().dim(),
errors['u - u_exact']))
# Compute convergence rates
from math import log as ln # log is a fenics name too
error_types = list(E[1][0].keys())
rates = {}
for degree in degrees:
rates[degree] = {}
for error_type in sorted(error_types):
rates[degree][error_type] = []
for i in range(num_meshes):
Ei = E[degree][i][error_type]
Eim1 = E[degree][i-1][error_type]
r = ln(Ei/Eim1)/ln(h[degree][i]/h[degree][i-1])
rates[degree][error_type].append(round(r,2))
return rates
Test problem
To demonstrate the computation of convergence rates, we will pick an exact solution \( \uex \) given by $$ \begin{equation*} \uex(x,y) = \sin(\omega\pi x)\sin(\omega\pi y) \end{equation*} $$ on the unit square. This choice implies \( f(x,y)=2\omega^2\pi^2 u(x,y) \). With \( \omega \) restricted to an integer, it follows that the boundary value is given by \( \ub=0 \).
We need to define the appropriate boundary conditions, the exact solution, and the \( f \) function in the code:
def boundary(x, on_boundary):
return on_boundary
bc = DirichletBC(V, Constant(0), boundary)
omega = 1.0
u_e = Expression('sin(omega*pi*x[0])*sin(omega*pi*x[1])', omega=omega)
f = 2*pi**2*omega**2*u_e
Experiments
An implementation of the computation of the convergence rate can be
found in the function convergence_rate_sin() in the demo program
ft11_poisson_bcs.py. We achieve some interesting results.
Using the error measure E5 based on the infinity norm of the
difference of the degrees of freedom, we obtain the following table.
| element | \( n=8\ \) | \( n=16\ \) | \( n=32\ \) | \( n=64\ \) | \( n=128 \) |
| \( \mathsf{P}_1 \) | 1.99 | 1.97 | 1.99 | 2.00 | 2.00 |
| \( \mathsf{P}_2 \) | 3.99 | 3.96 | 3.99 | 4.00 | 3.99 |
| \( \mathsf{P}_3 \) | 3.96 | 3.89 | 3.96 | 3.99 | 4.00 |
| \( \mathsf{P}_4 \) | 3.75 | 4.99 | 5.00 | 5.00 |
An entry like 3.96 for \( n=32 \) and \( \mathsf{P}_3 \) means that we estimate the rate 3.96 by comparing two meshes, with resolutions \( n=32 \) and \( n=16 \), using \( \mathsf{P}_3 \) elements. The coarsest mesh has \( n=4 \). The best estimates of the rates appear in the right-most column, since these rates are based on the finest resolutions and are hence deepest into the asymptotic regime.
The computations with \( \mathsf{P}_4 \) elements on a \( 2\times(128\times
128) \) mesh with a direct solver (UMFPACK) on a small laptop broke down
so these results have been left out of the table. Otherwise, we
achieve expected results: the error goes like \( h^{d+1} \) for elements
of degree \( d \). Also the \( L^2 \)-norm errors computed using the FEniCS
errornorm function show the
expected \( h^{d+1} \) rate for \( u \) and \( h^d \) for \( \nabla u \).
However, using (u_e - u)**2 for the error computation, with the same
degree for the interpolation of u_e as for u, gives strange
results:
| element | \( n=8\ \) | $n=16$\ | \( n=32\ \) | \( n=64\ \) | \( n=128 \) |
| \( \mathsf{P}_1 \) | 1.98 | 1.94 | 1.98 | 2.0 | 2.0 |
| \( \mathsf{P}_2 \) | 3.98 | 3.95 | 3.99 | 3.99 | 3.99 |
| \( \mathsf{P}_3 \) | 3.69 | 4.03 | 4.01 | 3.95 | 2.77 |
This is an example where it is important to interpolate u_e to a
higher-order space (polynomials of degree 3 are sufficient here) to
avoid computing a too optimistic convergence rate.
Checking convergence rates is the next best method for verifying PDE codes (the best being a numerical solution without approximation errors as in the section Examining the degrees of freedom and many other places in this tutorial).
Taking advantage of structured mesh data
Many readers have extensive experience with visualization and data analysis of 1D, 2D, and 3D scalar and vector fields on uniform, structured meshes, while FEniCS solvers exclusively work with unstructured meshes. Since it can many times be practical to work with structured data, we discuss in this section how to extract structured data for finite element solutions computed with FEniCS.
A necessary first step is to transform our Mesh object to an object
representing a rectangle with equally-shaped rectangular cells. The
second step is to transform the one-dimensional array of nodal values
to a two-dimensional array holding the values at the corners of the
cells in the structured mesh. We want to access a value by its \( i \) and
\( j \) indices, \( i \) counting cells in the \( x \) direction, and \( j \) counting
cells in the \( y \) direction. This transformation is in principle
straightforward, yet it frequently leads to obscure indexing errors,
so using software tools to ease the work is advantageous.
In the directory src/modules, associated with this book, we have
included the Python module BoxField that provides utilities for
working with structured mesh data in FEniCS. Given a finite element
function u, the following function returns a BoxField object that
represents u on a structured mesh:
def structured_mesh(u, divisions):
"""Represent u on a structured mesh."""
# u must have P1 elements, otherwise interpolate to P1 elements
u2 = u if u.ufl_element().degree() == 1 else \
interpolate(u, FunctionSpace(mesh, 'P', 1))
mesh = u.function_space().mesh()
from BoxField import fenics_function2BoxField
u_box = fenics_function2BoxField(
u2, mesh, divisions, uniform_mesh=True)
return u_box
(AL 8: Comment below needs to be modified if interpolation to P1 is included in BoxField.)
Note that we can only turn functions on meshes with \( \mathsf{P}_1 \) elements into
BoxField objects, so if u is based on another element type, we first
interpolate the scalar field onto a mesh with \( \mathsf{P}_1 \) elements. Also note
that to use the
function, we need to know the divisions into cells in the various
spatial directions (divisions).
The u_box object contains several useful data structures:
-
u_box.grid: object for the structured mesh -
u_box.grid.coor[X]: grid coordinates inX=0direction -
u_box.grid.coor[Y]: grid coordinates inY=1direction -
u_box.grid.coor[Z]: grid coordinates inZ=2direction -
u_box.grid.coorv[X]: vectorized version ofu_box.grid.coor[X](for vectorized computations or surface plotting) -
u_box.grid.coorv[Y]: vectorized version ofu_box.grid.coor[Y] -
u_box.grid.coorv[Z]: vectorized version ofu_box.grid.coor[Z] -
u_box.values:numpyarray holding theuvalues;u_box.values[i,j]holdsuat the mesh point with coordinates
(u_box.grid.coor[X], u_box.grid.coor[Y])
i and j indices?)
Iterating over points and values
Let us go back to the solver function in the
ft11_poisson_bcs.py code from
the section Postprocessing computations, compute u, map it onto a
BoxField object for a structured mesh representation, and
write out the coordinates and function values at all mesh points:
u = solver(p, f, u_b, nx, ny, 1, linear_solver='direct')
u_box = structured_mesh(u, (nx, ny))
u_ = u_box.values # numpy array
print('u_ is defined on a structured mesh with %s points' %
str(u_.shape))
# Iterate over 2D mesh points (i,j)
for j in range(u_.shape[1]):
for i in range(u_.shape[0]):
print('u[%d, %d] = u(%g, %g) = %g' %
(i, j,
u_box.grid.coor[X][i], u_box.grid.coor[X][j],
u_[i,j]))
Computing finite difference approximations
Using the multidimensional array u_ = u_box.values, we can easily
express finite difference approximations of derivatives:
x = u_box.grid.coor[X]
dx = x[1] - x[0]
u_xx = (u_[i - 1, j] - 2*u_[i, j] + u_[i + 1, j]) / dx**2
Making surface plots
The ability to access a finite element field as structured data is handy in many occasions, e.g., for visualization and data analysis. Using Matplotlib, we can create a surface plot, as shown in Figure 19 (upper left):
import matplotlib.pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
from matplotlib import cm
fig = plt.figure()
ax = fig.gca(projection='3d')
cv = u_box.grid.coorv # vectorized mesh coordinates
ax.plot_surface(cv[X], cv[Y], u_, cmap=cm.coolwarm,
rstride=1, cstride=1)
plt.title('Surface plot of solution')
The key issue is to know that the coordinates needed for the surface
plot is in u_box.grid.coorv and that the values are in u_.
Figure 19: Various plots of the solution on a structured mesh.
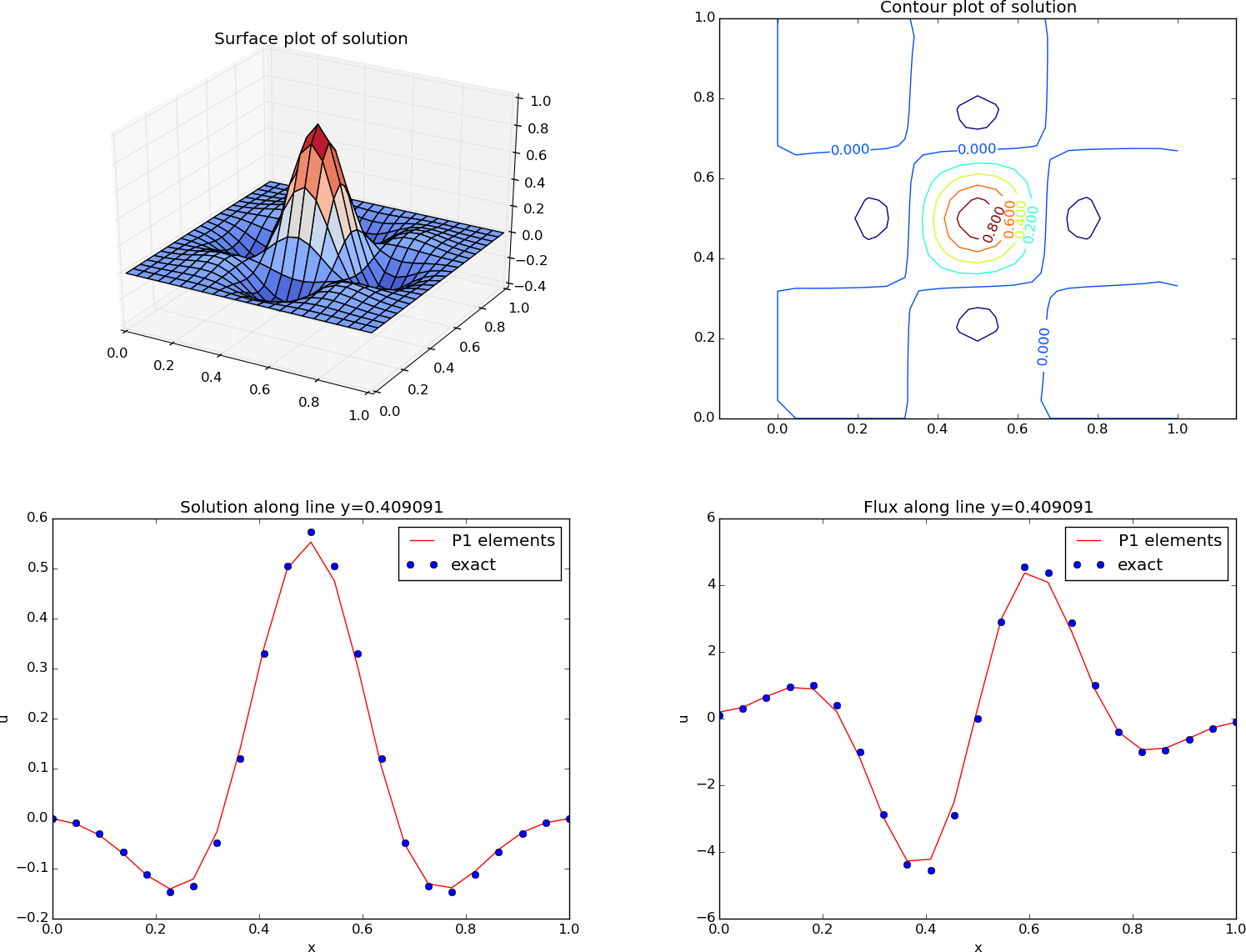
Making contour plots
A contour plot can also be made by Matplotlib:
fig = plt.figure()
ax = fig.gca()
levels = [1.5, 2.0, 2.5, 3.5]
cs = ax.contour(cv[X], cv[Y], u_, levels=levels)
plt.clabel(cs) # add labels to contour lines
plt.axis('equal')
plt.title('Contour plot of solution')
The result appears in Figure 19 (upper right).
Making curve plots through the domain
A handy feature of BoxField objects is the ability to give a starting
point in the domain and a direction, and then extract the field and
corresponding coordinates along the nearest line of mesh points. We have
already seen how to interpolate the solution along a line in the mesh, but
with BoxField you can pick out the computational points (vertices) for
examination of these points. Numerical methods often show improved behavior
at such points so this is of interest. For 3D fields
one can also extract data in a plane.
Say we want to plot \( u \) along
the line \( y=0.4 \). The mesh points, x, and the \( u \) values
along this line, u_val, can be extracted by
start = (0, 0.4)
x, u_val, y_fixed, snapped = u_box.gridline(start, direction=X)
The variable snapped is true if the line is snapped onto to nearst
gridline and in that case y_fixed holds the snapped
(altered) \( y \) value. (A keyword argument snap is by default True
to avoid interpolation and force snapping.)
A comparison of the numerical and exact solution along the line \( y\approx 0.41 \) (snapped from \( y=0.4 \)) is made by the following code:
start = (0, 0.4)
x, u_val, y_fixed, snapped = u_box.gridline(start, direction=X)
u_e_val = [u_b((x_, y_fixed)) for x_ in x]
plt.figure()
plt.plot(x, u_val, 'r-')
plt.plot(x, u_e_val, 'bo')
plt.legend(['P1 elements', 'exact'], loc='upper left')
plt.title('Solution along line y=%g' % y_fixed)
plt.xlabel('x'); plt.ylabel('u')
See Figure 19 (lower left) for the resulting curve plot.
Making curve plots of the flux
Let us also compare the numerical and exact flux \( -\kappa\partial u/\partial x \) along the same line as above:
flux_u = flux(u, p)
flux_u_x, flux_u_y = flux_u.split(deepcopy=True)
# Plot the numerical and exact flux along the same line
if flux_u_x.ufl_element().degree() == 1:
flux2_x = flux_u_x
else:
V1 = FunctionSpace(u.function_space().mesh(), 'P', 1)
flux2_x = interpolate(flux_x, V)
flux_u_x_box = structured_mesh(flux_u_x, (nx, ny))
x, flux_u_val, y_fixed, snapped = \
flux_u_x_box.gridline(start, direction=X)
y = y_fixed
plt.figure()
plt.plot(x, flux_u_val, 'r-')
plt.plot(x, flux_u_x_exact(x, y_fixed), 'bo')
plt.legend(['P1 elements', 'exact'], loc='upper right')
plt.title('Flux along line y=%g' % y_fixed)
plt.xlabel('x')
plt.ylabel('u')
The second plt.plot command
requires a function flux_u_x_exact(x,y) to be
available for the exact flux expression.
Note that Matplotlib is one choice of plotting package. With the unified interface in the SciTools package one can access Matplotlib, Gnuplot, MATLAB, OpenDX, VisIt, and other plotting engines through the same API.
Test problem
The graphics referred to in Figure 19 correspond to
a test problem with prescribed solution \( \uex = H(x)H(y) \), where
$$ H(x) = e^{-16(x-\frac{1}{2})^2}\sin(3\pi x)\tp$$
The corresponding right-hand side \( f \) is obtained by inserting the exact
solution into the PDE and differentiating as before.
Although it is easy to carry out the
differentiation of \( f \) by hand and hardcode the resulting expressions
in an Expression object, a more reliable habit is to use Python's
symbolic computing engine, SymPy, to perform mathematics and
automatically turn formulas into C++ syntax for Expression objects.
A short introduction was given in
the section FEniCS implementation.
We start out with defining the exact solution in sympy:
from sympy import exp, sin, pi # for use in math formulas
import sympy as sym
H = lambda x: exp(-16*(x-0.5)**2)*sin(3*pi*x)
x, y = sym.symbols('x[0], x[1]')
u = H(x)*H(y)
Turning the expression for u into C or C++ syntax for Expression objects
needs two steps. First we ask for the C code of the expression,
u_c = sym.printing.ccode(u)
Printing out u_c gives (the output is here manually broken into two
lines):
-exp(-16*pow(x[0] - 0.5, 2) - 16*pow(x[1] - 0.5, 2))*
sin(3*M_PI*x[0])*sin(3*M_PI*x[1])
The necessary syntax adjustment is replacing
the symbol M_PI for \( \pi \) in C/C++ by pi (or DOLFIN_PI):
u_c = u_c.replace('M_PI', 'pi')
u_b = Expression(u_c)
Thereafter, we can progress with the computation of \( f = -\nabla\cdot(\kappa\nabla u) \):
kappa = 1
f = sym.diff(-kappa*sym.diff(u, x), x) + \
sym.diff(-kappa*sym.diff(u, y), y)
f = sym.simplify(f)
f_c = sym.printing.ccode(f)
f_c = f_c.replace('M_PI', 'pi')
f = Expression(f_c)
We also need a Python function for the exact flux \( -\kappa\partial u/\partial x \):
flux_u_x_exact = sym.lambdify([x, y], -kappa*sym.diff(u, x),
modules='numpy')
It remains to define kappa = Constant(1) and set nx and ny before calling
solver to compute the finite element solution of this problem.