Implementation¶
This section presents the complete computational algorithm, its implementation in Python code, animation of the solution, and verification of the implementation.
A real implementation of the basic computational algorithm from the sections Formulating a recursive algorithm and Sketch of an implementation can be encapsulated in a function, taking all the input data for the problem as arguments. The physical input data consists of \(c\), \(I(x)\), \(V(x)\), \(f(x,t)\), \(L\), and \(T\). The numerical input is the mesh parameters \(\Delta t\) and \(\Delta x\).
Instead of specifying \(\Delta t\) and \(\Delta x\), we can specify one of them and the Courant number \(C\) instead, since having explicit control of the Courant number is convenient when investigating the numerical method. Many find it natural to prescribe the resolution of the spatial grid and set \(N_x\). The solver function can then compute \(\Delta t = CL/(cN_x)\). However, for comparing \(u(x,t)\) curves (as functions of \(x\)) for various Courant numbers it is more convenient to keep \(\Delta t\) fixed for all \(C\) and let \(\Delta x\) vary according to \(\Delta x = c\Delta t/C\). With \(\Delta t\) fixed, all frames correspond to the same time \(t\), and this simplifies animations that compare simulations with different mesh resolutions. Plotting functions of \(x\) with different spatial resolution is trivial, so it is easier to let \(\Delta x\) vary in the simulations than \(\Delta t\).
Callback function for user-specific actions¶
The solution at all spatial points at a new time level is stored in an
array u of length \(N_x+1\). We need to decide what do to with
this solution, e.g., visualize the curve, analyze the values, or write
the array to file for later use. The decision about what to do is left to
the user in the form of a user-suppled supplied function
user_action(u, x, t, n)
where u is the solution at the spatial points x at time t[n].
The user_action function is call from the solver at each time level n.
If the user wants to plot the solution or store the solution at a
time point, she needs to write such a function and take appropriate
actions inside it. We will show examples on many such user_action
functions.
Since the solver function make calls back to the user’s code via such a function, this type of function is called a callback function. When writing general software, like our solver function, which also needs to carry out special problem-dependent actions (like visualization), it is a common technique to leave those actions to user-supplied callback functions.
The solver function¶
A first attempt at a solver function is listed below.
import numpy as np
def solver(I, V, f, c, L, dt, C, T, user_action=None):
"""Solve u_tt=c^2*u_xx + f on (0,L)x(0,T]."""
Nt = int(round(T/dt))
t = np.linspace(0, Nt*dt, Nt+1) # Mesh points in time
dx = dt*c/float(C)
Nx = int(round(L/dx))
x = np.linspace(0, L, Nx+1) # Mesh points in space
C2 = C**2 # Help variable in the scheme
if f is None or f == 0 :
f = lambda x, t: 0
if V is None or V == 0:
V = lambda x: 0
u = np.zeros(Nx+1) # Solution array at new time level
u_1 = np.zeros(Nx+1) # Solution at 1 time level back
u_2 = np.zeros(Nx+1) # Solution at 2 time levels back
import time; t0 = time.clock() # for measuring CPU time
# Load initial condition into u_1
for i in range(0,Nx+1):
u_1[i] = I(x[i])
if user_action is not None:
user_action(u_1, x, t, 0)
# Special formula for first time step
n = 0
for i in range(1, Nx):
u[i] = u_1[i] + dt*V(x[i]) + \
0.5*C2*(u_1[i-1] - 2*u_1[i] + u_1[i+1]) + \
0.5*dt**2*f(x[i], t[n])
u[0] = 0; u[Nx] = 0
if user_action is not None:
user_action(u, x, t, 1)
# Switch variables before next step
u_2[:] = u_1; u_1[:] = u
for n in range(1, Nt):
# Update all inner points at time t[n+1]
for i in range(1, Nx):
u[i] = - u_2[i] + 2*u_1[i] + \
C2*(u_1[i-1] - 2*u_1[i] + u_1[i+1]) + \
dt**2*f(x[i], t[n])
# Insert boundary conditions
u[0] = 0; u[Nx] = 0
if user_action is not None:
if user_action(u, x, t, n+1):
break
# Switch variables before next step
u_2[:] = u_1; u_1[:] = u
cpu_time = t0 - time.clock()
return u, x, t, cpu_time
Verification: exact quadratic solution¶
We use the test problem derived in the section A slightly generalized model problem for verification. Below is a unit test based on this test problem and realized as a proper test function (compatible with the unit test frameworks nose or pytest).
def test_quadratic():
"""Check that u(x,t)=x(L-x)(1+t/2) is exactly reproduced."""
def u_exact(x, t):
return x*(L-x)*(1 + 0.5*t)
def I(x):
return u_exact(x, 0)
def V(x):
return 0.5*u_exact(x, 0)
def f(x, t):
return 2*(1 + 0.5*t)*c**2
L = 2.5
c = 1.5
C = 0.75
Nx = 6 # Very coarse mesh for this exact test
dt = C*(L/Nx)/c
T = 18
def assert_no_error(u, x, t, n):
u_e = u_exact(x, t[n])
diff = np.abs(u - u_e).max()
tol = 1E-13
assert diff < tol
solver(I, V, f, c, L, dt, C, T,
user_action=assert_no_error)
When this function resides in the file wave1D_u0.py, one can run ether
py.test or nosetests,
Terminal> py.test -s -v wave1D_u0.py
Terminal> nosetests -s -v wave1D_u0.py
to automatically run all test functions with name test_*().
Visualization: animating the solution¶
Now that we have verified the implementation it is time to do a
real computation where we also display the evolution of the waves
on the screen. Since the solver function knows nothing about
what type of visualizations we may want, it calls the callback function
user_action(u, x, t, n). We must therefore write this function and
find the proper statements for plotting the solution.
Function for administering the simulation¶
The following viz function
- defines a
user_actioncallback function for plotting the solution at each time level, - calls the
solverfunction, and - combines all the plots (in files) to video in different formats.
def viz(
I, V, f, c, L, dt, C, T, # PDE paramteres
umin, umax, # Interval for u in plots
animate=True, # Simulation with animation?
tool='matplotlib', # 'matplotlib' or 'scitools'
solver_function=solver, # Function with numerical algorithm
):
"""Run solver and visualize u at each time level."""
def plot_u_st(u, x, t, n):
"""user_action function for solver."""
plt.plot(x, u, 'r-',
xlabel='x', ylabel='u',
axis=[0, L, umin, umax],
title='t=%f' % t[n], show=True)
# Let the initial condition stay on the screen for 2
# seconds, else insert a pause of 0.2 s between each plot
time.sleep(2) if t[n] == 0 else time.sleep(0.2)
plt.savefig('frame_%04d.png' % n) # for movie making
class PlotMatplotlib:
def __call__(self, u, x, t, n):
"""user_action function for solver."""
if n == 0:
plt.ion()
self.lines = plt.plot(x, u, 'r-')
plt.xlabel('x'); plt.ylabel('u')
plt.axis([0, L, umin, umax])
plt.legend(['t=%f' % t[n]], loc='lower left')
else:
self.lines[0].set_ydata(u)
plt.legend(['t=%f' % t[n]], loc='lower left')
plt.draw()
time.sleep(2) if t[n] == 0 else time.sleep(0.2)
plt.savefig('tmp_%04d.png' % n) # for movie making
if tool == 'matplotlib':
import matplotlib.pyplot as plt
plot_u = PlotMatplotlib()
elif tool == 'scitools':
import scitools.std as plt # scitools.easyviz interface
plot_u = plot_u_st
import time, glob, os
# Clean up old movie frames
for filename in glob.glob('tmp_*.png'):
os.remove(filename)
# Call solver and do the simulaton
user_action = plot_u if animate else None
u, x, t, cpu = solver_function(
I, V, f, c, L, dt, C, T, user_action)
# Make video files
fps = 4 # frames per second
codec2ext = dict(flv='flv', libx264='mp4', libvpx='webm',
libtheora='ogg') # video formats
filespec = 'tmp_%04d.png'
movie_program = 'ffmpeg' # or 'avconv'
for codec in codec2ext:
ext = codec2ext[codec]
cmd = '%(movie_program)s -r %(fps)d -i %(filespec)s '\
'-vcodec %(codec)s movie.%(ext)s' % vars()
os.system(cmd)
if tool == 'scitools':
# Make an HTML play for showing the animation in a browser
plt.movie('tmp_*.png', encoder='html', fps=fps,
output_file='movie.html')
return cpu
Dissection of the code¶
The viz function can either use SciTools or Matplotlib for
visualizing the solution. The user_action function based on SciTools
is called plot_u_st, while the user_action function based on
Matplotlib is a bit more complicated as it is realized as a class and
needs statements that differ from those for making static plots.
SciTools can utilize both Matplotlib and Gnuplot (and many other
plotting programs) for doing the graphics, but Gnuplot is a relevant
choice for large \(N_x\) or in two-dimensional problems
as Gnuplot is significantly faster than
Matplotlib for screen animations.
A function inside another function, like plot_u_st in the above code
segment, has access to and remembers all the local variables in the
surrounding code inside the viz function (!). This is known in
computer science as a closure and is very convenient to program
with. For example, the plt and time modules defined outside
plot_u are accessible for plot_u_st when the function is called
(as user_action) in the solver function. Some may think, however,
that a class instead of a closure is a cleaner and
easier-to-understand implementation of the user action function, see
the section Building a general 1D wave equation solver.
The plot_u_st function just makes a standard SciTools plot command
for plotting u as a function of x at time t[n]. To achieve a
smooth animation, the plot command should take keyword arguments
instead of being broken into separate calls to xlabel, ylabel,
axis, time, and show. Several plot calls will automatically
cause an animation on the screen. In addition, we want to save each
frame in the animation to file. We then need a filename where the
frame number is padded with zeros, here tmp_0000.png,
tmp_0001.png, and so on. The proper printf construction is then
tmp_%04d.png.
The solver is called with an argument plot_u as user_function.
If the user chooses to use SciTools, plot_u is the plot_u_st
callback function, but for Matplotlib it is an instance of the
class PlotMatplotlib. Also this class makes use of variables
defined in the viz function: plt and time.
With Matplotlib, one has to make the first plot the standard way, and
then update the \(y\) data in the plot at every time level. The update
requires active use of the returned value from plt.plot in the first
plot. This value would need to be stored in a local variable if we
were to use a closure for the user_action function when doing the
animation with Matplotlib. It is much easier to store the
variable as a class attribute self.lines. Since the class is essentially a
function, we implement the function as the special method __call__
such that the instance plot_u(u, x, t, n) can be called as a standard
callback function from solver.
Making movie files¶
From the
frame_*.png files containing the frames in the animation we can
make video files.
We use the ffmpeg (or avconv) program to combine individual
plot files to movies in modern formats: Flash, MP4, Webm, and Ogg.
A typical ffmpeg (or avconv) command for creating a movie file
in Ogg format
with 4 frames per second built from a collection of
plot files with names generated by frame_%04d.png,
look like
Terminal> ffmpeg -r 4 -i frame_%04d.png -c:v libtheora movie.ogg
The different formats require different video encoders (-c:v) to
be installed: Flash applies flv, WebM applies libvpx, and MP4
applies libx264:
Terminal> ffmpeg -r 4 -i frame_%04d.png -c:v flv movie.flv
Terminal> ffmpeg -r 4 -i frame_%04d.png -c:v libvpx movie.webm
Terminal> ffmpeg -r 4 -i frame_%04d.png -c:v libx264 movie.mp4
Players like vlc, mplayer, gxine, and totem
can be used to play these movie files.
Note that padding the frame counter with zeros in the frame_*.png
files, as specified by the %04d format, is essential so that the wildcard
notation frame_*.png expands to the correct set of files.
The viz function creates a ffmpeg or avconv command
with the proper arguments for each of the formats Flash, MP4, WebM,
and Ogg. The task is greatly simplified by having a
codec2ext dictionary for mapping
video codec names to filename extensions.
Only
two formats are actually needed to ensure that all browsers can
successfully play the video: MP4 and WebM.
Some animations consisting of a large number of plot files may not
be properly combined into a video using ffmpeg or avconv.
A method that always works is to play the PNG files as an animation
in a browser using JavaScript code in an HTML file.
The SciTools package has a function movie (or a stand-alone command
scitools movie) for creating such an HTML player. The plt.movie
call in the viz function shows how the function is used.
The file movie.html can be loaded into a browser and features
a user interface where the speed of the animation can be controlled.
Note that the movie in this case consists of the movie.html file
and all the frame files tmp_*.png.
Skipping frames for animation speed¶
Sometimes the time step is small and \(T\) is large, leading to an
inconveniently large number of plot files and a slow animation on the
screen. The solution to such a problem is to decide on a total number
of frames in the animation, num_frames, and plot the solution only for
every skip_frame frames. For example, setting skip_frame=5 leads
to plots of every 5 frames. The default value skip_frame=1 plots
every frame.
The total number of time levels (i.e., maximum
possible number of frames) is the length of t, t.size (or len(t)),
so if we want num_frames frames in the animation,
we need to plot every t.size/num_frames frames:
skip_frame = int(t.size/float(num_frames))
if n % skip_frame == 0 or n == t.size-1:
st.plot(x, u, 'r-', ...)
The initial condition (n=0) included by n % skip_frame == 0,
as well as every skip_frame-th frame.
As n % skip_frame == 0 will very seldom be true for the
very final frame, we must also check if n == t.size-1 to
get the final frame included.
A simple choice of numbers may illustrate the formulas: say we have
801 frames in total (t.size) and we allow only 60 frames to be
plotted. Then we need to plot every 801/60 frame, which with integer
division yields 13 as every. Using the mod function, n % every,
this operation is zero every time n can be divided by 13 without a
remainder. That is, the if test is true when n equals \(0, 13, 26,
39, ..., 780, 801\). The associated code is included in the plot_u
function in the file wave1D_u0v.py.
Running a case¶
The first demo of our 1D wave equation solver concerns vibrations of a string that is initially deformed to a triangular shape, like when picking a guitar string:
We choose \(L=75\) cm, \(x_0=0.8L\), \(a=5\) mm, and a time frequency \(\nu = 440\) Hz. The relation between the wave speed \(c\) and \(\nu\) is \(c=\nu\lambda\), where \(\lambda\) is the wavelength, taken as \(2L\) because the longest wave on the string form half a wavelength. There is no external force, so \(f=0\), and the string is at rest initially so that \(V=0\).
Regarding numerical parameters, we need to specify a \(\Delta t\).
Sometimes it is more natural to think of a spatial resolution instead
of a time step. A natural semi-coarse spatial resolution in the present
problem is \(N_x=50\). We can then choose the associated \(\Delta t\) (as required
by the viz and solver functions) as the stability limit:
\(\Delta t = L/(N_xc)\). This is the \(\Delta t\) to be specified,
but notice that if \(C<1\), the actual \(\Delta x\) computed in solver gets
larger than \(L/N_x\): \(\Delta x = c\Delta t/C = L/(N_xC)\). (The reason
is that we fix \(\Delta t\) and adjust \(\Delta x\), so if \(C\) gets
smaller, the code implements this effect in terms of a larger \(\Delta x\).)
A function for setting the physical and numerical parameters and
calling viz in this application goes as follows:
def guitar(C):
"""Triangular wave (pulled guitar string)."""
L = 0.75
x0 = 0.8*L
a = 0.005
freq = 440
wavelength = 2*L
c = freq*wavelength
omega = 2*pi*freq
num_periods = 1
T = 2*pi/omega*num_periods
# Choose dt the same as the stability limit for Nx=50
dt = L/50./c
def I(x):
return a*x/x0 if x < x0 else a/(L-x0)*(L-x)
umin = -1.2*a; umax = -umin
cpu = viz(I, 0, 0, c, L, dt, C, T, umin, umax,
animate=True, tool='scitools')
The associated program has the name wave1D_u0.py. Run the program and watch the movie of the vibrating string.
Working with a scaled PDE model¶
Depending on the model, it may be a substantial job to establish consistent and relevant physical parameter values for a case. The guitar string example illustrates the point. However, by scaling the mathematical problem we can often reduce the need to estimate physical parameters dramatically. The scaling technique consists of introducing new independent and dependent variables, with the aim that the absolute value of these is not very large or small, but preferably around unity in size. We introduce the dimensionless variables
Here, \(L\) is a typical length scale, e.g., the length of the domain, and \(a\) is a typical size of \(u\), e.g., determined from the initial condition: \(a=\max_x|I(x)|\).
Inserting these new variables in the PDE and noting that
by the chain rule, one gets
in case \(f=0\). Dropping the bars, we arrive at the scaled PDE
which has not parameter \(c^2\) anymore. The initial conditions are scaled as
and
resulting in
In the common case \(V=0\) we see that there are no physical parameters to be estimated in the PDE model!
If we have a program implemented for the physical wave equation with
dimensions, we can obtain the dimensionless, scaled version by
setting \(c=1\). The initial condition of a guitar string,
given in (33), gets its scaled form by choosing
\(a=1\), \(L=1\), and \(x_0\in [0,1]\). This means that we only need to
decide on the \(x_0\) value as a fraction of unity, because
the scaled problem corresponds to setting all
other parameters to unity. In the code we can just set
a=c=L=1, x0=0.8, and there is no need to calculate with
wavelengths and frequencies to estimate \(c\)!
The only non-trivial parameter to estimate in the scaled problem is the final end time of the simulation, or more precisely, how it relates to periods in periodic solutions in time, since we often want to express the end time as a certain number of periods. The period in the dimensionless problem is 2, so the end time can be set to the desired number of periods times 2.
Why the dimensionless period is 2 can be explained by the following reasoning. Suppose as \(u\) behaves as \(\cos (\omega t)\) in time in variables with dimension. The corresponding period is then \(P=2\pi/\omega\), but we need to estimate \(\omega\). A typical solution of the wave equation is \(u(x,t)=A\cos(kx)\cos(\omega t)\), where \(A\) is an amplitude and \(k\) is related to the wave length \(\lambda\) in space: \(\lambda = 2\pi/k\). Both \(\lambda\) and \(A\) will be given by the initial condition \(I(x)\). Inserting this \(u(x,t)\) in the PDE yields \(-\omega^2 = -c^2k^2\), i.e., \(\omega = kc\). The period is therefore \(P=2\pi/(kc)\). If the boundary conditions are \(u(0,t)=u(0,L)\), we need to have \(kL = n\pi\) for integer \(n\). The period becomes \(P=2L/nc\). The longest period is \(P=2L/c\). The dimensionless period is \(\tilde P\) is obtained by dividing \(P\) by the time scale \(L/c\), which results in \(\tilde P=2\). Shorter waves in the initial condition will have a dimensionless shorter period \(\tilde P=2/n\) (\(n>1\)).
Vectorization¶
The computational algorithm for solving the wave equation visits one mesh point at a time and evaluates a formula for the new value \(u_i^{n+1}\) at that point. Technically, this is implemented by a loop over array elements in a program. Such loops may run slowly in Python (and similar interpreted languages such as R and MATLAB). One technique for speeding up loops is to perform operations on entire arrays instead of working with one element at a time. This is referred to as vectorization, vector computing, or array computing. Operations on whole arrays are possible if the computations involving each element is independent of each other and therefore can, at least in principle, be performed simultaneously. Vectorization not only speeds up the code on serial computers, but also makes it easy to exploit parallel computing.
Operations on slices of arrays¶
Efficient computing with numpy arrays demands that we avoid loops
and compute with entire arrays at once (or at least large portions of them).
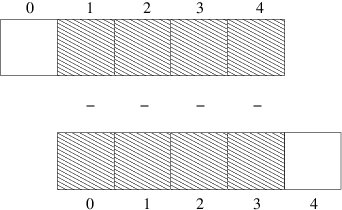
Consider this calculation of differences \(d_i = u_{i+1}-u_i\):
n = u.size
for i in range(0, n-1):
d[i] = u[i+1] - u[i]
All the differences here are independent of each other.
The computation of d can therefore alternatively be done by
subtracting the array \((u_0,u_1,\ldots,u_{n-1})\) from
the array where the elements are shifted one index upwards:
\((u_1,u_2,\ldots,u_n)\), see Figure Illustration of subtracting two slices of two arrays.
The former subset of the array can be
expressed by u[0:n-1],
u[0:-1], or just
u[:-1], meaning from index 0 up to,
but not including, the last element (-1). The latter subset
is obtained by u[1:n] or u[1:],
meaning from index 1 and the rest of the array.
The computation of d can now be done without an explicit Python loop:
d = u[1:] - u[:-1]
or with explicit limits if desired:
d = u[1:n] - u[0:n-1]
Indices with a colon, going from an index to (but not including) another
index are called slices. With numpy arrays, the computations
are still done by loops, but in efficient, compiled, highly optimized
C or Fortran code. Such loops are sometimes referred to as vectorized
loops. Such loops can also easily be distributed
among many processors on parallel computers. We say that the scalar code
above, working on an element (a scalar) at a time, has been replaced by
an equivalent vectorized code. The process of vectorizing code is called
vectorization.
Test your understanding
Newcomers to vectorization are encouraged to choose
a small array u, say with five elements,
and simulate with pen and paper
both the loop version and the vectorized version above.
Finite difference schemes basically contain differences between array elements with shifted indices. As an example, consider the updating formula
for i in range(1, n-1):
u2[i] = u[i-1] - 2*u[i] + u[i+1]
The vectorization consists of replacing the loop by arithmetics on
slices of arrays of length n-2:
u2 = u[:-2] - 2*u[1:-1] + u[2:]
u2 = u[0:n-2] - 2*u[1:n-1] + u[2:n] # alternative
Note that the length of u2 becomes n-2. If u2 is already an array of
length n and we want to use the formula to update all the “inner”
elements of u2, as we will when solving a 1D wave equation, we can write
u2[1:-1] = u[:-2] - 2*u[1:-1] + u[2:]
u2[1:n-1] = u[0:n-2] - 2*u[1:n-1] + u[2:n] # alternative
The first expression’s right-hand side is realized by the
following steps, involving temporary arrays with intermediate results,
since each array operation can only involve one or two arrays.
The numpy package performs the first line above in
four steps:
temp1 = 2*u[1:-1]
temp2 = u[:-2] - temp1
temp3 = temp2 + u[2:]
u2[1:-1] = temp3
We need three temporary arrays, but a user does not need to worry about such temporary arrays.
Common mistakes with array slices
Array expressions with slices demand that the slices have the same shape. It easy to make a mistake in, e.g.,
u2[1:n-1] = u[0:n-2] - 2*u[1:n-1] + u[2:n]
and write
u2[1:n-1] = u[0:n-2] - 2*u[1:n-1] + u[1:n]
Now u[1:n] has wrong length (n-1) compared to the other array
slices, causing a ValueError and the message
could not broadcast input array from shape 103 into shape 104
(if n is 105). When such errors occur one must closely examine
all the slices. Usually, it is easier to get upper limits of slices
right when they use -1 or -2 or empty limit rather than
expressions involving the length.
Another common mistake is to forget the slice in the array on the left-hand side,
u2 = u[0:n-2] - 2*u[1:n-1] + u[1:n]
This is really crucial: now u2 becomes a new array of length
n-2, which is the wrong length as we have no entries for the boundary
values. We meant to insert the right-hand side array into the
in the original u2 array for the entries that correspond to the
internal points in the mesh (1:n-1 or 1:-1).
Vectorization may also work nicely with functions. To illustrate, we may extend the previous example as follows:
def f(x):
return x**2 + 1
for i in range(1, n-1):
u2[i] = u[i-1] - 2*u[i] + u[i+1] + f(x[i])
Assuming u2, u, and x all have length n, the vectorized
version becomes
u2[1:-1] = u[:-2] - 2*u[1:-1] + u[2:] + f(x[1:-1])
Obviously, f must be able to take an array as argument for f[x[1:-1])
to make sense.
Finite difference schemes expressed as slices¶
We now have the necessary tools to vectorize the wave equation algorithm as described mathematically in the section Formulating a recursive algorithm and through code in the section The solver function. There are three loops: one for the initial condition, one for the first time step, and finally the loop that is repeated for all subsequent time levels. Since only the latter is repeated a potentially large number of times, we limit our vectorization efforts to this loop:
for i in range(1, Nx):
u[i] = 2*u_1[i] - u_2[i] + \
C2*(u_1[i-1] - 2*u_1[i] + u_1[i+1])
The vectorized version becomes
u[1:-1] = - u_2[1:-1] + 2*u_1[1:-1] + \
C2*(u_1[:-2] - 2*u_1[1:-1] + u_1[2:])
or
u[1:Nx] = 2*u_1[1:Nx]- u_2[1:Nx] + \
C2*(u_1[0:Nx-1] - 2*u_1[1:Nx] + u_1[2:Nx+1])
The program
wave1D_u0v.py
contains a new version of the function solver where both the scalar
and the vectorized loops are included (the argument version is
set to scalar or vectorized, respectively).
Verification¶
We may reuse the quadratic solution \({u_{\small\mbox{e}}}(x,t)=x(L-x)(1+{\frac{1}{2}}t)\) for
verifying also the vectorized code. A test function can now verify
both the scalar and the vectorized version. Moreover, we may
use a user_action function that compares the computed and exact
solution at each time level and performs a test:
def test_quadratic():
"""
Check the scalar and vectorized versions work for
a quadratic u(x,t)=x(L-x)(1+t/2) that is exactly reproduced.
"""
# The following function must work for x as array or scalar
u_exact = lambda x, t: x*(L - x)*(1 + 0.5*t)
I = lambda x: u_exact(x, 0)
V = lambda x: 0.5*u_exact(x, 0)
# f is a scalar (zeros_like(x) works for scalar x too)
f = lambda x, t: np.zeros_like(x) + 2*c**2*(1 + 0.5*t)
L = 2.5
c = 1.5
C = 0.75
Nx = 3 # Very coarse mesh for this exact test
dt = C*(L/Nx)/c
T = 18
def assert_no_error(u, x, t, n):
u_e = u_exact(x, t[n])
tol = 1E-13
diff = np.abs(u - u_e).max()
assert diff < tol
solver(I, V, f, c, L, dt, C, T,
user_action=assert_no_error, version='scalar')
solver(I, V, f, c, L, dt, C, T,
user_action=assert_no_error, version='vectorized')
Lambda functions
The code segment above demonstrates how to achieve very compact code, without degraded readability, by use of lambda functions for the various input parameters that require a Python function. In essence,
f = lambda x, t: L*(x-t)**2
is equivalent to
def f(x, t):
return L(x-t)**2
Note that lambda functions can just contain a single expression and no statements.
One advantage with lambda functions is that they can be used directly in calls:
solver(I=lambda x: sin(pi*x/L), V=0, f=0, ...)
Efficiency measurements¶
The wave1D_u0v.py contains our new solver function with both
scalar and vectorized code. For comparing the efficiency
of scalar versus vectorized code, we need a viz function
as discussed in the section Visualization: animating the solution.
All of this viz function can be reused, except the call
to solver_function. This call lacks the parameter
version, which we want to set to vectorized and scalar
for our efficiency measurements.
One solution is to copy the viz code from wave1D_u0 into
wave1D_u0v.py and add a version argument to the solver_function call.
Taking into account how much quite complicated animation code we
then duplicate, this is not a good idea.
Introducing the version argument in wave1D_u0.viz is not
a good solution since version has no meaning in that file.
Solution 1¶
Calling viz in wave1D_u0 with solver_function as our new
solver in wave1D_u0v works fine, since this solver has
version='vectorized' as default value. The problem arises when we
want to test version='vectorized'. The simplest solution is then
to use wave1D_u0.solver instead. We make a new viz function
in wave1D_u0v.py that has a version argument and that just
calls wave1D_u0.viz:
def viz(
I, V, f, c, L, dt, C, T, # PDE paramteres
umin, umax, # Interval for u in plots
animate=True, # Simulation with animation?
tool='matplotlib', # 'matplotlib' or 'scitools'
solver_function=solver, # Function with numerical algorithm
version='vectorized', # 'scalar' or 'vectorized'
):
import wave1D_u0
if version == 'vectorized':
# Reuse viz from wave1D_u0, but with the present
# modules' new vectorized solver (which has
# version='vectorized' as default argument;
# wave1D_u0.viz does not feature this argument)
cpu = wave1D_u0.viz(
I, V, f, c, L, dt, C, T, umin, umax,
animate, tool, solver_function=solver)
elif version == 'scalar':
# Call wave1D_u0.viz with a solver with
# scalar code and use wave1D_u0.solver.
cpu = wave1D_u0.viz(
I, V, f, c, L, dt, C, T, umin, umax,
animate, tool,
solver_function=wave1D_u0.solver)
Solution 2¶
There is a more advanced, fancier solution featuring a very useful trick:
we can make a new function that will always call wave1D_u0v.solver
with version='scalar'. The functools.partial function from
standard Python takes a function func as argument and
a series of positional and keyword arguments and returns a
new function that will call func with the supplied arguments,
while the user can control all the other arguments in func.
Consider a trivial example,
def f(a, b, c=2):
return a + b + c
We want to ensure that f is always called with c=3, i.e., f
has only two “free” arguments a and b.
This functionality is obtained by
import functools
f2 = functools.partial(f, c=3)
print f2(1, 2) # results in 1+2+3=6
Now f2 calls f with whatever the user supplies as a and b,
but c is always 3.
Back to our viz code, we can do
import functools
# Call scalar with version fixed to `scalar`
scalar_solver = functools.partial(scalar, version='scalar')
cpu = wave1D_u0.viz(
I, V, f, c, L, dt, C, T, umin, umax,
animate, tool, solver_function=scalar_solver)
The new scalar_solver takes the same arguments as
wave1D_u0.scalar and calls wave1D_u0v.scalar,
but always supplies the extra argument
version='scalar'. When sending this solver_function
to wave1D_u0.viz, the latter will call wave1D_u0v.solver
with all the I, V, f, etc., arguments we supply, plus
version='scalar'.
Efficiency experiments¶
We now have a viz function that can call our solver function both in
scalar and vectorized mode. The function run_efficiency_experiments
in wave1D_u0v.py performs a set of experiments and reports the
CPU time spent in the scalar and vectorized solver for
the previous string vibration example with spatial mesh resolutions
\(N_x=50,100,200,400,800\). Running this function reveals
that the vectorized
code runs substantially faster: the vectorized code runs approximately
\(N_x/10\) times as fast as the scalar code!
Remark on the updating of arrays¶
At the end of each time step we need to update the u_2 and u_1
arrays such that they have the right content for the next time step:
u_2[:] = u_1
u_1[:] = u
The order here is important! (Updating u_1 first, makes u_2 equal
to u, which is wrong.)
The assignment u_1[:] = u copies the content of the u array into
the elements of the u_1 array. Such copying takes time, but
that time is negligible compared to the time needed for
computing u from the finite difference formula,
even when the formula has a vectorized implementation.
However, efficiency of program code is a key topic when solving
PDEs numerically (particularly when there are two or three
space dimensions), so it must be mentioned that there exists a
much more efficient way of making the arrays u_2 and u_1
ready for the next time step. The idea is based on switching
references and explained as follows.
A Python variable is actually a reference to some object (C programmers
may think of pointers). Instead of copying data, we can let u_2
refer to the u_1 object and u_1 refer to the u object.
This is a very efficiency operation (like switching pointers in C).
A naive implementation like
u_2 = u_1
u_1 = u
will fail, however, because now u_2 refers to the u_1 object,
but then the name u_1 refers to u, so that this u object
has two references, u_1 and u, while our third array, originally
referred to by u_2 has no more references and is lost.
This means that the variables u, u_1, and u_2 refer to two
arrays and not three. Consequently, the computations at the next
time level will be messed up since updating the elements in
u will imply updating the elements in u_1 too so the solution
at the previous time step, which is crucial in our formulas, is
destroyed.
While u_2 = u_1 is fine, u_1 = u is problematic, so
the solution to this problem is to ensure that u
points to the u_2 array. This is mathematically wrong, but
new correct values will be filled into u at the next time step
and make it right.
The correct switch of references is
tmp = u_2
u_2 = u_1
u_1 = u
u = tmp
We can get rid of the temporary reference tmp by writing
u_2, u_1, u = u_1, u, u_2
This switching of references for updating our arrays will be used in later implementations.
Caution
The update u_2, u_1, u = u_1, u, u_2 leaves wrong content in u
at the final time step. This means that if we return u, as we
do in the example codes here, we actually return u_2, which is
obviously wrong. It is therefore important to adjust the content
of u to u = u_1 before returning u.
Exercises¶
Exercise 1: Simulate a standing wave¶
The purpose of this exercise is to simulate standing waves on \([0,L]\) and illustrate the error in the simulation. Standing waves arise from an initial condition
where \(m\) is an integer and \(A\) is a freely chosen amplitude. The corresponding exact solution can be computed and reads
a) Explain that for a function \(\sin kx\cos \omega t\) the wave length in space is \(\lambda = 2\pi /k\) and the period in time is \(P=2\pi/\omega\). Use these expressions to find the wave length in space and period in time of \({u_{\small\mbox{e}}}\) above.
b)
Import the solver function wave1D_u0.py into a new file
where the viz function is reimplemented such that it
plots either the numerical and the exact solution, or the error.
c) Make animations where you illustrate how the error \(e^n_i ={u_{\small\mbox{e}}}(x_i, t_n)- u^n_i\) develops and increases in time. Also make animations of \(u\) and \({u_{\small\mbox{e}}}\) simultaneously.
Hint 1. Quite long time simulations are needed in order to display significant discrepancies between the numerical and exact solution.
Hint 2. A possible set of parameters is \(L=12\), \(m=9\), \(c=2\), \(A=1\), \(N_x=80\), \(C=0.8\). The error mesh function \(e^n\) can be simulated for 10 periods, while 20-30 periods are needed to show significant differences between the curves for the numerical and exact solution.
Filename: wave_standing.
Remarks¶
The important parameters for numerical quality are \(C\) and \(k\Delta x\), where \(C=c\Delta t/\Delta x\) is the Courant number and \(k\) is defined above (\(k\Delta x\) is proportional to how many mesh points we have per wave length in space, see the section Numerical dispersion relation for explanation).
Exercise 2: Add storage of solution in a user action function¶
Extend the plot_u function in the file wave1D_u0.py to also store
the solutions u in a list.
To this end, declare all_u as
an empty list in the viz function, outside plot_u, and perform
an append operation inside the plot_u function. Note that a
function, like plot_u, inside another function, like viz,
remembers all local variables in viz function, including all_u,
even when plot_u is called (as user_action) in the solver function.
Test both all_u.append(u) and all_u.append(u.copy()).
Why does one of these constructions fail to store the solution correctly?
Let the viz function return the all_u list
converted to a two-dimensional numpy array.
Filename: wave1D_u0_s_store.
Exercise 3: Use a class for the user action function¶
Redo Exercise 2: Add storage of solution in a user action function using a class for the
user action function. That is, define a class Action where
the all_u list is an attribute, and implement the user action
function as a method (the special method __call__ is a natural
choice). The class versions avoids that the user action function
depends on parameters defined outside the function (such as all_u
in Exercise 2: Add storage of solution in a user action function).
Filename: wave1D_u0_s2c.
Exercise 4: Compare several Courant numbers in one movie¶
The goal of this exercise is to make movies where several curves,
corresponding to different Courant numbers, are visualized.
Import the solver function from the wave1D_u0_s movie
in a new file wave_compare.py. Reimplement the viz function
such that it can take a list of C values as argument
and create a movie with solutions corresponding to the given C
values. The plot_u function must be changed to store the solution
in an array (see Exercise 2: Add storage of solution in a user action function or
Exercise 3: Use a class for the user action function for details), solver must be
computed for each value of the Courant number, and finally
one must run through each time step and plot all the spatial
solution curves in one figure and store it in a file.
The challenge in such a visualization is to ensure that the curves in
one plot corresponds to the same time point. The easiest remedy is to
keep the time and space resolution constant and change the wave
velocity \(c\) to change the Courant number.
Filename: wave_numerics_comparison.
Project 5: Calculus with 1D mesh functions¶
This project explores integration and differentiation of mesh functions, both with scalar and vectorized implementations. We are given a mesh function \(f_i\) on a spatial one-dimensional mesh \(x_i=i\Delta x\), \(i=0,\ldots,N_x\), over the interval \([a,b]\).
a) Define the discrete derivative of \(f_i\) by using centered differences at internal mesh points and one-sided differences at the end points. Implement a scalar version of the computation in a Python function and write an associated unit test for the linear case \(f(x)=4x-2.5\) where the discrete derivative should be exact.
b) Vectorize the implementation of the discrete derivative. Extend the unit test to check the validity of the implementation.
c) To compute the discrete integral \(F_i\) of \(f_i\), we assume that the mesh function \(f_i\) varies linearly between the mesh points. Let \(f(x)\) be such a linear interpolant of \(f_i\). We then have
The exact integral of a piecewise linear function \(f(x)\) is given by the Trapezoidal rule. S how that if \(F_{i}\) is already computed, we can find \(F_{i+1}\) from
Make a function for the scalar implementation of the discrete integral as a mesh function. That is, the function should return \(F_i\) for \(i=0,\ldots,N_x\). For a unit test one can use the fact that the above defined discrete integral of a linear function (say \(f(x)=4x-2.5\)) is exact.
d) Vectorize the implementation of the discrete integral. Extend the unit test to check the validity of the implementation.
Hint.
Interpret the recursive formula for \(F_{i+1}\) as a sum.
Make an array with each element of the sum and use the “cumsum”
(numpy.cumsum) operation to compute the accumulative sum:
numpy.cumsum([1,3,5]) is [1,4,9].
e)
Create a class MeshCalculus that can integrate and differentiate
mesh functions. The class can just define some methods that call
the previously implemented Python functions. Here is an example
on the usage:
import numpy as np
calc = MeshCalculus(vectorized=True)
x = np.linspace(0, 1, 11) # mesh
f = np.exp(x) # mesh function
df = calc.differentiate(f, x) # discrete derivative
F = calc.integrate(f, x) # discrete anti-derivative
Filename: mesh_calculus_1D.