The discrete solution
- The numerical solution is a mesh function: \( u_i^n \approx \uex(x_i,t_n) \)
- Finite difference stencil (or scheme): equation for \( u^n_i \) involving neighboring space-time points
Waves on a string can be modeled by the wave equation
$$ \frac{\partial^2 u}{\partial t^2} = c^2 \frac{\partial^2 u}{\partial x^2} $$
\( u(x,t) \) is the displacement of the string
$$
\begin{align}
\frac{\partial^2 u}{\partial t^2} &=
c^2 \frac{\partial^2 u}{\partial x^2}, \quad &x\in (0,L),\ t\in (0,T]
\tag{1}\\
u(x,0) &= I(x), \quad &x\in [0,L]
\tag{2}\\
\frac{\partial}{\partial t}u(x,0) &= 0, \quad &x\in [0,L]
\tag{3}\\
u(0,t) & = 0, \quad &t\in (0,T]
\tag{4}\\
u(L,t) & = 0, \quad &t\in (0,T]
\tag{5}
\end{align}
$$
Rule for number of initial and boundary conditions:
Ooops!
Mesh in time:
$$
\begin{equation}
0 = t_0 < t_1 < t_2 < \cdots < t_{N_t-1} < t_{N_t} = T \end{equation}
$$
Mesh in space:
$$
\begin{equation}
0 = x_0 < x_1 < x_2 < \cdots < x_{N_x-1} < x_{N_x} = L \end{equation}
$$
Uniform mesh with constant mesh spacings \( \Delta t \) and \( \Delta x \):
$$
\begin{equation}
x_i = i\Delta x,\ i=0,\ldots,N_x,\quad
t_i = n\Delta t,\ n=0,\ldots,N_t
\end{equation}
$$

Let the PDE be satisfied at all interior mesh points:
$$
\begin{equation}
\frac{\partial^2}{\partial t^2} u(x_i, t_n) =
c^2\frac{\partial^2}{\partial x^2} u(x_i, t_n),
\tag{6}
\end{equation}
$$
for \( i=1,\ldots,N_x-1 \) and \( n=1,\ldots,N_t-1 \).
For \( n=0 \) we have the initial conditions \( u=I(x) \) and \( u_t=0 \), and at the boundaries \( i=0,N_x \) we have the boundary condition \( u=0 \).
Widely used finite difference formula for the second-order derivative:
$$ \frac{\partial^2}{\partial t^2}u(x_i,t_n)\approx
\frac{u_i^{n+1} - 2u_i^n + u^{n-1}_i}{\Delta t^2}= [D_tD_t u]^n_i$$
and
$$ \frac{\partial^2}{\partial x^2}u(x_i,t_n)\approx
\frac{u_{i+1}^{n} - 2u_i^n + u^{n}_{i-1}}{\Delta x^2} = [D_xD_x u]^n_i
$$
Replace derivatives by differences:
$$
\begin{equation}
\frac{u_i^{n+1} - 2u_i^n + u^{n-1}_i}{\Delta t^2} =
c^2\frac{u_{i+1}^{n} - 2u_i^n + u^{n}_{i-1}}{\Delta x^2},
\tag{7}
\end{equation}
$$
In operator notation:
$$
\begin{equation}
[D_tD_t u = c^2 D_xD_x]^{n}_i
\tag{8}
\end{equation}
$$
$$ [D_{2t} u]^n_i = 0,\quad n=0\quad\Rightarrow\quad
u^{n-1}_i=u^{n+1}_i,\quad i=0,\ldots,N_x$$
The other initial condition \( u(x,0)=I(x) \) can be computed by
$$ u_i^0 = I(x_i),\quad i=0,\ldots,N_x$$
Write out \( [D_tD_t u = c^2 D_xD_x]^{n}_i \) and solve for \( u^{n+1}_i \),
$$
\begin{equation}
u^{n+1}_i = -u^{n-1}_i + 2u^n_i + C^2
\left(u^{n}_{i+1}-2u^{n}_{i} + u^{n}_{i-1}\right)
\tag{9}
\end{equation}
$$
$$
\begin{equation}
C = c\frac{\Delta t}{\Delta x},
\end{equation}
$$
is known as the (dimensionless) Courant number

Initial condition:
$$ [D_{2t}u=0]^0_i\quad\Rightarrow\quad u^{-1}_i=u^1_i$$
Insert in stencil \( [D_tD_tu = c^2D_xD_x]^0_i \) to get
$$
\begin{equation}
u_i^1 = u^0_i - \half
C^2\left(u^{n}_{i+1}-2u^{n}_{i} + u^{n}_{i-1}\right)
\tag{10}
\end{equation}
$$
web page or a movie file.
u[i] stores \( u^{n+1}_i \)u_1[i] stores \( u^n_i \)u_2[i] stores \( u^{n-1}_i \)
u is the unknown to be computed (a spatial mesh
function), u_k is the computed spatial mesh function k
time steps back in time.
# Given mesh points as arrays x and t (x[i], t[n])
dx = x[1] - x[0]
dt = t[1] - t[0]
C = c*dt/dx # Courant number
Nt = len(t)-1
C2 = C**2 # Help variable in the scheme
# Set initial condition u(x,0) = I(x)
for i in range(0, Nx+1):
u_1[i] = I(x[i])
# Apply special formula for first step, incorporating du/dt=0
for i in range(1, Nx):
u[i] = u_1[i] - 0.5*C**2(u_1[i+1] - 2*u_1[i] + u_1[i-1])
u[0] = 0; u[Nx] = 0 # Enforce boundary conditions
# Switch variables before next step
u_2[:], u_1[:] = u_1, u
for n in range(1, Nt):
# Update all inner mesh points at time t[n+1]
for i in range(1, Nx):
u[i] = 2u_1[i] - u_2[i] - \
C**2(u_1[i+1] - 2*u_1[i] + u_1[i-1])
# Insert boundary conditions
u[0] = 0; u[Nx] = 0
# Switch variables before next step
u_2[:], u_1[:] = u_1, u
Add source term \( f \) and nonzero initial condition \( u_t(x,0) \):
$$
\begin{align}
u_{tt} &= c^2 u_{xx} + f(x,t),
\tag{11}\\
u(x,0) &= I(x), \quad &x\in [0,L]
\tag{12}\\
u_t(x,0) &= V(x), \quad &x\in [0,L]
\tag{13}\\
u(0,t) & = 0, \quad & t>0,
\tag{14}\\
u(L,t) & = 0, \quad &t>0
\tag{15}
\end{align}
$$
$$
\begin{equation}
[D_tD_t u = c^2 D_xD_x + f]^{n}_i
\tag{16}
\end{equation}
$$
Writing out and solving for the unknown \( u^{n+1}_i \):
$$
\begin{equation}
u^{n+1}_i = -u^{n-1}_i + 2u^n_i + C^2
(u^{n}_{i+1}-2u^{n}_{i} + u^{n}_{i-1}) + \Delta t^2 f^n_i
\tag{17}
\end{equation}
$$
Centered difference for \( u_t(x,0) = V(x) \):
$$ [D_{2t}u = V]^0_i\quad\Rightarrow\quad u^{-1}_i = u^{1}_i - 2\Delta t V_i,$$
Inserting this in the stencil (17) for \( n=0 \) leads to
$$
\begin{equation}
u^{1}_i = u^0_i - \Delta t V_i + {\half}
C^2
\left(u^{n}_{i+1}-2u^{n}_{i} + u^{n}_{i-1}\right) + \half\Delta t^2 f^n_i
\tag{18}
\end{equation}
$$
$$
\begin{equation}
\uex(x,y,t)) = A\sin\left(\frac{\pi}{L}x\right)
\cos\left(\frac{\pi}{L}ct\right)
\tag{19}
\end{equation}
$$
$$ \uex(x,t) = x(L-x)\sin t$$
PDE \( u_{tt}=c^2u_{xx}+f \):
$$ -x(L-x)\sin t = -2\sin t + f\quad\Rightarrow f = (2 - x(L-x))\sin t$$
Initial conditions become
$$
\begin{align*}
u(x,0) &= I(x) = 0\\
u_t(x,0) &= V(x) = (2 - x(L-x))\cos t
\end{align*}
$$
Boundary conditions:
$$ u(x,0) = u(x,L) = 0 $$
Here, choose \( \uex \) such that \( \uex(x,0)=\uex(L,0)=0 \):
$$ \uex (x,t) = x(L-x)(1+{\half}t), $$
Insert in the PDE and find \( f \):
$$ f(x,t)=2(1+t)c^2$$
Initial conditions:
$$ I(x) = x(L-x),\quad V(x)={\half}x(L-x) $$
We want to show that \( \uex \) also solves the discrete equations!
Useful preliminary result:
$$
\begin{align}
\lbrack D_tD_t t^2\rbrack^n &= \frac{t_{n+1}^2 - 2t_n^2 + t_{n-1}^2}{\Delta t^2}
= (n+1)^2 -n^2 + (n-1)^2 = 2\\
\lbrack D_tD_t t\rbrack^n &= \frac{t_{n+1} - 2t_n + t_{n-1}}{\Delta t^2}
= \frac{((n+1) -n + (n-1))\Delta t}{\Delta t^2} = 0
\end{align}
$$
Hence,
$$ [D_tD_t \uex]^n_i = x_i(L-x_i)[D_tD_t (1+{\half}t)]^n =
x_i(L-x_i){\half}[D_tD_t t]^n = 0$$
$$
\begin{align*}
\lbrack D_xD_x \uex\rbrack^n_i &=
(1+{\half}t_n)\lbrack D_xD_x (xL-x^2)\rbrack_i =
(1+{\half}t_n)\lbrack LD_xD_x x - D_xD_x x^2\rbrack_i \\
&= -2(1+{\half}t_n)
\end{align*}
$$
Now, \( f^n_i = 2(1+{\half}t_n)c^2 \) and we get
$$ [D_tD_t \uex - c^2D_xD_x\uex - f]^n_i = 0 - c^2(-1)2(1 + {\half}t_n
+ 2(1+{\half}t_n)c^2 = 0$$
Moreover, \( \uex(x_i,0)=I(x_i) \), \( \partial \uex/\partial t = V(x_i) \) at \( t=0 \), and \( \uex(x_0,t)=\uex(x_{N_x},0)=0 \). Also the modified scheme for the first time step is fulfilled by \( \uex(x_i,t_n) \).
Later we show that the exact solution of the discrete equations can be obtained by \( C=1 \) (!)
def user_action(u, x, t, n):
# u[i] at spatial mesh points x[i] at time t[n]
# plot u
# or store u
from numpy import *
def solver(I, V, f, c, L, Nx, C, T, user_action=None):
"""Solve u_tt=c^2*u_xx + f on (0,L)x(0,T]."""
x = linspace(0, L, Nx+1) # Mesh points in space
dx = x[1] - x[0]
dt = C*dx/c
Nt = int(round(T/dt))
t = linspace(0, Nt*dt, Nt+1) # Mesh points in time
C2 = C**2 # Help variable in the scheme
if f is None or f == 0 :
f = lambda x, t: 0
if V is None or V == 0:
V = lambda x: 0
u = zeros(Nx+1) # Solution array at new time level
u_1 = zeros(Nx+1) # Solution at 1 time level back
u_2 = zeros(Nx+1) # Solution at 2 time levels back
import time; t0 = time.clock() # for measuring CPU time
# Load initial condition into u_1
for i in range(0,Nx+1):
u_1[i] = I(x[i])
if user_action is not None:
user_action(u_1, x, t, 0)
# Special formula for first time step
n = 0
for i in range(1, Nx):
u[i] = u_1[i] + dt*V(x[i]) + \
0.5*C2*(u_1[i-1] - 2*u_1[i] + u_1[i+1]) + \
0.5*dt**2*f(x[i], t[n])
u[0] = 0; u[Nx] = 0
if user_action is not None:
user_action(u, x, t, 1)
# Switch variables before next step
u_2[:], u_1[:] = u_1, u
for n in range(1, Nt):
# Update all inner points at time t[n+1]
for i in range(1, Nx):
u[i] = - u_2[i] + 2*u_1[i] + \
C2*(u_1[i-1] - 2*u_1[i] + u_1[i+1]) + \
dt**2*f(x[i], t[n])
# Insert boundary conditions
u[0] = 0; u[Nx] = 0
if user_action is not None:
if user_action(u, x, t, n+1):
break
# Switch variables before next step
u_2[:], u_1[:] = u_1, u
cpu_time = t0 - time.clock()
return u, x, t, cpu_time
Exact solution of the PDE problem and the discrete equations: \( \uex (x,t) = x(L-x)(1+{\half}t) \)
import nose.tools as nt
def test_quadratic():
"""Check that u(x,t)=x(L-x)(1+t/2) is exactly reproduced."""
def exact_solution(x, t):
return x*(L-x)*(1 + 0.5*t)
def I(x):
return exact_solution(x, 0)
def V(x):
return 0.5*exact_solution(x, 0)
def f(x, t):
return 2*(1 + 0.5*t)*c**2
L = 2.5
c = 1.5
Nx = 3 # Very coarse mesh
C = 0.75
T = 18
u, x, t, cpu = solver(I, V, f, c, L, Nx, C, T)
u_e = exact_solution(x, t[-1])
diff = abs(u - u_e).max()
nt.assert_almost_equal(diff, 0, places=14)
Make a viz function for animating the curve, with plotting
in a user_action function plot_u:
def viz(I, V, f, c, L, Nx, C, T, umin, umax, animate=True):
"""Run solver and visualize u at each time level."""
import scitools.std as plt
import time, glob, os
def plot_u(u, x, t, n):
"""user_action function for solver."""
plt.plot(x, u, 'r-',
xlabel='x', ylabel='u',
axis=[0, L, umin, umax],
title='t=%f' % t[n], show=True)
# Let the initial condition stay on the screen for 2
# seconds, else insert a pause of 0.2 s between each plot
time.sleep(2) if t[n] == 0 else time.sleep(0.2)
plt.savefig('frame_%04d.png' % n) # for movie making
# Clean up old movie frames
for filename in glob.glob('frame_*.png'):
os.remove(filename)
user_action = plot_u if animate else None
u, x, t, cpu = solver(I, V, f, c, L, Nx, C, T, user_action)
# Make movie files
fps = 4 # Frames per second
plt.movie('frame_*.png', encoder='html', fps=fps,
output_file='movie.html')
codec2ext = dict(flv='flv', libx64='mp4', libvpx='webm',
libtheora='ogg')
filespec = 'frame_%04d.png'
movie_program = 'avconv' # or 'ffmpeg'
for codec in codec2ext:
ext = codec2ext[codec]
cmd = '%(movie_program)s -r %(fps)d -i %(filespec)s '\
'-vcodec %(codec)s movie.%(ext)s' % vars()
os.system(cmd)
Note: plot_u is function inside function and remembers the
local variables in viz (known as a closure).
'something_%04d.png' % frame_counter
Terminal> scitools movie encoder=html output_file=movie.html \
fps=4 frame_*.png # web page with a player
Terminal> avconv -r 4 -i frame_%04d.png -vcodec flv movie.flv
Terminal> avconv -r 4 -i frame_%04d.png -vcodec libtheora movie.ogg
Terminal> avconv -r 4 -i frame_%04d.png -vcodec libx264 movie.mp4
Terminal> avconv -r 4 -i frame_%04d.png -vcodec libtheora movie.ogg
Terminal> avconv -r 4 -i frame_%04d.png -vcodec libpvx movie.webm
%04d) is essential for correct sequence of
frames in something_*.png (Unix alphanumeric sort)frame_*.png files before making a new movie
$$
\begin{equation}
I(x) = \left\lbrace
\begin{array}{ll}
ax/x_0, & x < x_0\\
a(L-x)/(L-x_0), & \hbox{otherwise}
\end{array}\right.
\tag{20}
\end{equation}
$$
Appropriate data:
def guitar(C):
"""Triangular wave (pulled guitar string)."""
L = 0.75
x0 = 0.8*L
a = 0.005
freq = 440
wavelength = 2*L
c = freq*wavelength
omega = 2*pi*freq
num_periods = 1
T = 2*pi/omega*num_periods
Nx = 50
def I(x):
return a*x/x0 if x < x0 else a/(L-x0)*(L-x)
umin = -1.2*a; umax = -umin
cpu = viz(I, 0, 0, c, L, Nx, C, T, umin, umax, animate=True)
Program: wave1D_u0_s.py.
Introduce new \( x \), \( t \), and \( u \) without dimension:
$$ \bar x = \frac{x}{L},\quad \bar t = \frac{c}{L}t,\quad
\bar u = \frac{u}{a}
$$
Insert this in the PDE (with \( f=0 \)) and dropping bars
$$ u_{tt} = u_{xx}$$
Initial condition: set \( a=1 \), \( L=1 \), and \( x_0\in [0,1] \) in (20).
In the code: set a=c=L=1, x0=0.8, and there is no need to calculate with
wavelengths and frequencies to estimate \( c \)!
Just one challenge: determine the period of the waves and an appropriate end time (see the text for details).
numpy) code instead of explicit loopsNext: vectorized loops
n = u.size
for i in range(0, n-1):
d[i] = u[i+1] - u[i]
numpy code: u[1:n] - u[0:n-1] or just u[1:] - u[:-1]
Newcomers to vectorization are encouraged to choose
a small array u, say with five elements,
and simulate with pen and paper
both the loop version and the vectorized version.
Finite difference schemes basically contains differences between array elements with shifted indices. Consider the updating formula
for i in range(1, n-1):
u2[i] = u[i-1] - 2*u[i] + u[i+1]
The vectorization consists of replacing the loop by arithmetics on
slices of arrays of length n-2:
u2 = u[:-2] - 2*u[1:-1] + u[2:]
u2 = u[0:n-2] - 2*u[1:n-1] + u[2:n] # alternative
Note: u2 gets length n-2.
If u2 is already an array of length n, do update on "inner" elements
u2[1:-1] = u[:-2] - 2*u[1:-1] + u[2:]
u2[1:n-1] = u[0:n-2] - 2*u[1:n-1] + u[2:n] # alternative
Include a function evaluation too:
def f(x):
return x**2 + 1
# Scalar version
for i in range(1, n-1):
u2[i] = u[i-1] - 2*u[i] + u[i+1] + f(x[i])
# Vectorized version
u2[1:-1] = u[:-2] - 2*u[1:-1] + u[2:] + f(x[1:-1])
Scalar loop:
for i in range(1, Nx):
u[i] = 2*u_1[i] - u_2[i] + \
C2*(u_1[i-1] - 2*u_1[i] + u_1[i+1])
Vectorized loop:
u[1:-1] = - u_2[1:-1] + 2*u_1[1:-1] + \
C2*(u_1[:-2] - 2*u_1[1:-1] + u_1[2:])
or
u[1:Nx] = 2*u_1[1:Nx]- u_2[1:Nx] + \
C2*(u_1[0:Nx-1] - 2*u_1[1:Nx] + u_1[2:Nx+1])
Program: wave1D_u0_sv.py
def test_quadratic():
"""
Check the scalar and vectorized versions work for
a quadratic u(x,t)=x(L-x)(1+t/2) that is exactly reproduced.
"""
# The following function must work for x as array or scalar
exact_solution = lambda x, t: x*(L - x)*(1 + 0.5*t)
I = lambda x: exact_solution(x, 0)
V = lambda x: 0.5*exact_solution(x, 0)
# f is a scalar (zeros_like(x) works for scalar x too)
f = lambda x, t: zeros_like(x) + 2*c**2*(1 + 0.5*t)
L = 2.5
c = 1.5
Nx = 3 # Very coarse mesh
C = 1
T = 18 # Long time integration
def assert_no_error(u, x, t, n):
u_e = exact_solution(x, t[n])
diff = abs(u - u_e).max()
nt.assert_almost_equal(diff, 0, places=13)
solver(I, V, f, c, L, Nx, C, T,
user_action=assert_no_error, version='scalar')
solver(I, V, f, c, L, Nx, C, T,
user_action=assert_no_error, version='vectorized')
Note:
wave1D_u0_sv.py for \( N_x \) as 50,100,200,400,800
and measuring the CPU timeMuch bigger improvements for 2D and 3D codes!
$$
\begin{equation}
\frac{\partial u}{\partial n} \equiv \normalvec\cdot\nabla u = 0
\tag{21}
\end{equation}
$$
For a 1D domain \( [0,L] \):
$$
\left.\frac{\partial}{\partial n}\right\vert_{x=L} =
\frac{\partial}{\partial x},\quad
\left.\frac{\partial}{\partial n}\right\vert_{x=0} = -
\frac{\partial}{\partial x}
$$
Boundary condition terminology:
$$
\begin{equation}
\frac{u_{-1}^n - u_1^n}{2\Delta x} = 0
\tag{22}
\end{equation}
$$
$$
\frac{u_{-1}^n - u_1^n}{2\Delta x} = 0
$$
$$
\begin{equation}
u^{n+1}_i = -u^{n-1}_i + 2u^n_i + 2C^2
\left(u^{n}_{i+1}-u^{n}_{i}\right),\quad i=0 \end{equation}
$$
Discrete equation for computing \( u^3_0 \) in terms of \( u^2_0 \), \( u^1_0 \), and \( u^2_1 \):
Animation in a web page or a movie file.
i = 0
ip1 = i+1
im1 = ip1 # i-1 -> i+1
u[i] = u_1[i] + C2*(u_1[im1] - 2*u_1[i] + u_1[ip1])
i = Nx
im1 = i-1
ip1 = im1 # i+1 -> i-1
u[i] = u_1[i] + C2*(u_1[im1] - 2*u_1[i] + u_1[ip1])
# Or just one loop over all points
for i in range(0, Nx+1):
ip1 = i+1 if i < Nx else i-1
im1 = i-1 if i > 0 else i+1
u[i] = u_1[i] + C2*(u_1[im1] - 2*u_1[i] + u_1[ip1])
Program wave1D_dn0.py
web page or a movie file.
| Notation | Python |
| \( \Ix \) | Ix |
| \( \setb{\Ix} \) | Ix[0] |
| \( \sete{\Ix} \) | Ix[-1] |
| \( \setl{\Ix} \) | Ix[1:] |
| \( \setr{\Ix} \) | Ix[:-1] |
| \( \seti{\Ix} \) | Ix[1:-1] |
Index sets for a problem in the \( x,t \) plane:
$$
\begin{equation}
\Ix = \{0,\ldots,N_x\},\quad \It = \{0,\ldots,N_t\},
\end{equation}
$$
Implemented in Python as
Ix = range(0, Nx+1)
It = range(0, Nt+1)
A finite difference scheme can with the index set notation be specified as
$$
\begin{align*}
u^{n+1}_i &= -u^{n-1}_i + 2u^n_i + C^2
\left(u^{n}_{i+1}-2u^{n}_{i}+u^{n}_{i-1}\right),
\quad i\in\seti{\Ix},\ n\in\seti{\It}\\
u_i &= 0,
\quad i=\setb{\Ix},\ n\in\seti{\It}\\
u_i &= 0,
\quad i=\sete{\Ix},\ n\in\seti{\It}
\end{align*}
$$
Corresponding implementation:
for n in It[1:-1]:
for i in Ix[1:-1]:
u[i] = - u_2[i] + 2*u_1[i] + \
C2*(u_1[i-1] - 2*u_1[i] + u_1[i+1])
i = Ix[0]; u[i] = 0
i = Ix[-1]; u[i] = 0
Program wave1D_dn.py
Add ghost points:
u = zeros(Nx+3)
u_1 = zeros(Nx+3)
u_2 = zeros(Nx+3)
x = linspace(0, L, Nx+1) # Mesh points without ghost points
u[-1] will always mean the last element in u0,..,Nx+2
u = zeros(Nx+3)
Ix = range(1, u.shape[0]-1)
# Boundary values: u[Ix[0]], u[Ix[-1]]
# Set initial conditions
for i in Ix:
u_1[i] = I(x[i-Ix[0]]) # Note i-Ix[0]
# Loop over all physical mesh points
for i in Ix:
u[i] = - u_2[i] + 2*u_1[i] + \
C2*(u_1[i-1] - 2*u_1[i] + u_1[i+1])
# Update ghost values
i = Ix[0] # x=0 boundary
u[i-1] = u[i+1]
i = Ix[-1] # x=L boundary
u[i-1] = u[i+1]
Program: wave1D_dn0_ghost.py.
Heterogeneous media: varying \( c=c(x) \)
$$
\begin{equation}
\frac{\partial^2 u}{\partial t^2} =
\frac{\partial}{\partial x}\left( q(x)
\frac{\partial u}{\partial x}\right) + f(x,t)
\tag{23}
\end{equation}
$$
This equation sampled at a mesh point \( (x_i,t_n) \):
$$
\frac{\partial^2 }{\partial t^2} u(x_i,t_n) =
\frac{\partial}{\partial x}\left( q(x_i)
\frac{\partial}{\partial x} u(x_i,t_n)\right) + f(x_i,t_n),
$$
The principal idea is to first discretize the outer derivative.
Define
$$ \phi = q(x)
\frac{\partial u}{\partial x}
$$
Then use a centered derivative around \( x=x_i \) for the derivative of \( \phi \):
$$
\left[\frac{\partial\phi}{\partial x}\right]^n_i
\approx \frac{\phi_{i+\half} - \phi_{i-\half}}{\Delta x}
= [D_x\phi]^n_i
$$
Then discretize the inner operators:
$$
\phi_{i+\half} = q_{i+\half}
\left[\frac{\partial u}{\partial x}\right]^n_{i+\half}
\approx q_{i+\half} \frac{u^n_{i+1} - u^n_{i}}{\Delta x}
= [q D_x u]_{i+\half}^n
$$
Similarly,
$$
\phi_{i-\half} = q_{i-\half}
\left[\frac{\partial u}{\partial x}\right]^n_{i-\half}
\approx q_{i-\half} \frac{u^n_{i} - u^n_{i-1}}{\Delta x}
= [q D_x u]_{i-\half}^n
$$
These intermediate results are now combined to
$$
\begin{equation}
\left[
\frac{\partial}{\partial x}\left( q(x)
\frac{\partial u}{\partial x}\right)\right]^n_i
\approx \frac{1}{\Delta x^2}
\left( q_{i+\half} \left({u^n_{i+1} - u^n_{i}}\right)
- q_{i-\half} \left({u^n_{i} - u^n_{i-1}}\right)\right)
\tag{24}
\end{equation}
$$
In operator notation:
$$
\begin{equation}
\left[
\frac{\partial}{\partial x}\left( q(x)
\frac{\partial u}{\partial x}\right)\right]^n_i
\approx [D_xq D_x u]^n_i
\tag{25}
\end{equation}
$$
$$
\begin{align}
q_{i+\half} &\approx
\half\left( q_{i} + q_{i+1}\right) =
[\overline{q}^{x}]_i
\quad &\hbox{(arithmetic mean)}
\tag{26}\\
q_{i+\half} &\approx
2\left( \frac{1}{q_{i}} + \frac{1}{q_{i+1}}\right)^{-1}
\quad &\hbox{(harmonic mean)}
\tag{27}\\
q_{i+\half} &\approx
\left(q_{i}q_{i+1}\right)^{1/2}
\quad &\hbox{(geometric mean)}
\tag{28}
\end{align}
$$
The arithmetic mean in (26) is by far the most used averaging technique.
$$
\begin{equation}
\lbrack D_tD_t u = D_x\overline{q}^{x}D_x u + f\rbrack^{n}_i
\tag{29}
\end{equation}
$$
We clearly see the type of finite differences and averaging!
Write out and solve wrt \( u_i^{n+1} \):
$$
\begin{align}
u^{n+1}_i &= - u_i^{n-1} + 2u_i^n + \left(\frac{\Delta x}{\Delta t}\right)^2\times \nonumber\\
&\quad \left(
\half(q_{i} + q_{i+1})(u_{i+1}^n - u_{i}^n) -
\half(q_{i} + q_{i-1})(u_{i}^n - u_{i-1}^n)\right)
+ \nonumber\\
& \quad \Delta t^2 f^n_i
\tag{30}
\end{align}
$$
Consider \( \partial u/\partial x=0 \) at \( x=L=N_x\Delta x \):
$$ \frac{u_{i+1}^{n} - u_{i-1}^n}{2\Delta x} = 0\quad u_{i+1}^n = u_{i-1}^n,
\quad i=N_x
$$
Insert \( u_{i+1}^n=u_{i-1}^n \) in the stencil (30) for \( i=N_x \) and obtain
$$
u^{n+1}_i \approx
- u_i^{n-1} + 2u_i^n + \left(\frac{\Delta x}{\Delta t}\right)^2
2q_{i}(u_{i-1}^n - u_{i}^n) + \Delta t^2 f^n_i
$$
(We have used \( q_{i+\half} + q_{i-\half}\approx 2q_i \).)
Alternative: assume \( dq/dx=0 \) (simpler).
Assume c[i] holds \( c_i \) the spatial mesh points
for i in range(1, Nx):
u[i] = - u_2[i] + 2*u_1[i] + \
C2*(0.5*(q[i] + q[i+1])*(u_1[i+1] - u_1[i]) - \
0.5*(q[i] + q[i-1])*(u_1[i] - u_1[i-1])) + \
dt2*f(x[i], t[n])
Here: C2=(dt/dx)**2
Vectorized version:
u[1:-1] = - u_2[1:-1] + 2*u_1[1:-1] + \
C2*(0.5*(q[1:-1] + q[2:])*(u_1[2:] - u_1[1:-1]) -
0.5*(q[1:-1] + q[:-2])*(u_1[1:-1] - u_1[:-2])) + \
dt2*f(x[1:-1], t[n])
Neumann condition \( u_x=0 \): same ideas as in 1D (modified stencil or ghost cells).
$$
\begin{equation}
\varrho(x)\frac{\partial^2 u}{\partial t^2} =
\frac{\partial}{\partial x}\left( q(x)
\frac{\partial u}{\partial x}\right) + f(x,t)
\tag{31}
\end{equation}
$$
A natural scheme is
$$
\begin{equation}
[\varrho D_tD_t u = D_x\overline{q}^xD_x u + f]^n_i
\end{equation}
$$
Or
$$
\begin{equation}
[D_tD_t u = \varrho^{-1}D_x\overline{q}^xD_x u + f]^n_i
\end{equation}
$$
No need to average \( \varrho \), just sample at \( i \)
Why do waves die out?
Simplest damping model (for physical behavior, see demo):
$$
\begin{equation}
\frac{\partial^2 u}{\partial t^2} + \color{red}{b\frac{\partial u}{\partial t}}
= c^2\frac{\partial^2 u}{\partial x^2} + f(x,t),
\tag{32}
\end{equation}
$$
\( b \geq 0 \): prescribed damping coefficient.
Discretization via centered differences to ensure \( \Oof{\Delta t^2} \) error:
$$
\begin{equation}
[D_tD_t u + bD_{2t}u = c^2D_xD_x u + f]^n_i
\tag{33}
\end{equation}
$$
Need special formula for \( u^1_i \) + special stencil (or ghost cells) for Neumann conditions.
The program wave1D_dn_vc.py solves a fairly general 1D wave equation:
$$
\begin{align}
u_t &= (c^2(x)u_x)_x + f(x,t),\quad &x\in (0,L),\ t\in (0,T]
\tag{34}\\
u(x,0) &= I(x),\quad &x\in [0,L]
\tag{35}\\
u_t(x,0) &= V(t),\quad &x\in [0,L]
\tag{36}\\
u(0,t) &= U_0(t)\hbox{ or } u_x(0,t)=0,\quad &t\in (0,T]
\tag{37}\\
u(L,t) &= U_L(t)\hbox{ or } u_x(L,t)=0,\quad &t\in (0,T]
\tag{38}
\end{align}
$$
Can be adapted to many needs.
The function pulse in wave1D_dn_vc.py offers four
initial conditions:
gaussian)Can locate the initial pulse at \( x=0 \) or in the middle
>>> import wave1D_dn_vc as w
>>> w.pulse(loc='left', pulse_tp='cosinehat', Nx=50, every_frame=10)
Constant wave velocity \( c \):
$$
\begin{equation}
\frac{\partial^2 u}{\partial t^2} = c^2\nabla^2 u\hbox{ for }\xpoint\in\Omega\subset\Real^d,\ t\in (0,T]
\tag{39}
\end{equation}
$$
Variable wave velocity:
$$
\begin{equation}
\varrho\frac{\partial^2 u}{\partial t^2} = \nabla\cdot (q\nabla u) + f\hbox{ for }\xpoint\in\Omega\subset\Real^d,\ t\in (0,T]
\tag{40}
\end{equation}
$$
3D, constant \( c \):
$$
\begin{equation*} \nabla^2 u = \frac{\partial^2 u}{\partial x^2} +
\frac{\partial^2 u}{\partial y^2} + \frac{\partial^2 u}{\partial z^2}
\end{equation*}
$$
2D, variable \( c \):
$$
\begin{equation}
\varrho(x,y)
\frac{\partial^2 u}{\partial t^2} =
\frac{\partial}{\partial x}\left( q(x,y)
\frac{\partial u}{\partial x}\right)
+
\frac{\partial}{\partial y}\left( q(x,y)
\frac{\partial u}{\partial y}\right)
+ f(x,y,t)
\end{equation}
$$
Compact notation:
$$
\begin{align}
u_{tt} &= c^2(u_{xx} + u_{yy} + u_{zz}) + f,
\tag{41}\\
\varrho u_{tt} &= (q u_x)_x + (q u_z)_z + (q u_z)_z + f
\tag{42}
\end{align}
$$
We need one boundary condition at each point on \( \partial\Omega \):
PDEs with second-order time derivative need two initial conditions:
$$
[D_tD_t u = c^2(D_xD_x u + D_yD_yu) + f]^n_{i,j,k},
$$
Written out in detail:
$$
\begin{align*}
\frac{u^{n+1}_{i,j} - 2u^{n}_{i,j} + u^{n-1}_{i,j}}{\Delta t^2}
&= c^2
\frac{u^{n}_{i+1,j} - 2u^{n}_{i,j} + u^{n}_{i-1,j}}{\Delta x^2}
+ \nonumber\\
&\quad c^2\frac{u^{n}_{i,j+1} - 2u^{n}_{i,j} + u^{n}_{i,j-1}}{\Delta y^2}
+ f^n_{i,j},
\end{align*}
$$
\( u^{n-1}_{i,j} \) and \( u^n_{i,j} \) are known, solve for \( u^{n+1}_{i,j} \):
$$ u^{n+1}_{i,j} = 2u^n_{i,j} + u^{n-1}_{i,j} + c^2\Delta t^2[D_xD_x u + D_yD_y u]^n_{i,j}$$
$$ [D_{2t}u = V]^0_{i,j}\quad\Rightarrow\quad u^{-1}_{i,j} = u^1_{i,j} - 2\Delta t V_{i,j}
$$
$$ u^{n+1}_{i,j} = u^n_{i,j} -2\Delta V_{i,j} + {\half}
c^2\Delta t^2[D_xD_x u + D_yD_y u]^n_{i,j}$$
3D wave equation:
$$ \varrho u_{tt} = (qu_x)_x + (qu_y)_y + (qu_z)_z + f(x,y,z,t) $$
Just apply the 1D discretization for each term:
$$
\begin{equation}
[\varrho D_tD_t u = (D_x\overline{q}^x D_x u +
D_y\overline{q}^y D_yu + D_z\overline{q}^z D_z u) + f]^n_{i,j,k}
\end{equation}
$$
Need special formula for \( u^1_{i,j,k} \) (use \( [D_{2t}u=V]^0 \) and stencil for \( n=0 \)).
Written out:
$$
\begin{align*}
u^{n+1}_{i,j,k} &= - u^{n-1}_{i,j,k} + 2u^{n}_{i,j,k} + \\
&= \frac{1}{\varrho_{i,j,k}}\frac{1}{\Delta x^2} ( \half(q_{i,j,k} + q_{i+1,j,k})(u^{n}_{i+1,j,k} - u^{n}_{i,j,k}) - \\
&\qquad\quad \half(q_{i-1,j,k} + q_{i,j,k})(u^{n}_{i,j,k} - u^{n}_{i-1,j,k})) + \\
&= \frac{1}{\varrho_{i,j,k}}\frac{1}{\Delta x^2} ( \half(q_{i,j,k} + q_{i,j+1,k})(u^{n}_{i,j+1,k} - u^{n}_{i,j,k}) - \\
&\qquad\quad\half(q_{i,j-1,k} + q_{i,j,k})(u^{n}_{i,j,k} - u^{n}_{i,j-1,k})) + \\
&= \frac{1}{\varrho_{i,j,k}}\frac{1}{\Delta x^2} ( \half(q_{i,j,k} + q_{i,j,k+1})(u^{n}_{i,j,k+1} - u^{n}_{i,j,k}) -\\
&\qquad\quad \half(q_{i,j,k-1} + q_{i,j,k})(u^{n}_{i,j,k} - u^{n}_{i,j,k-1})) + \\
+ &\qquad \Delta t^2 f^n_{i,j,k}
\end{align*}
$$
Use ideas from 1D! Example: \( \frac{\partial u}{\partial n} \) at \( y=0 \), \( \frac{\partial u}{\partial n} = -\frac{\partial u}{\partial y} \)
Boundary condition discretization:
$$ [-D_{2y} u = 0]^n_{i,0}\quad\Rightarrow\quad \frac{u^n_{i,1}-u^n_{i,-1}}{2\Delta y} = 0,\ i\in\Ix
$$
Insert \( u^n_{i,-1}=u^n_{i,1} \) in the stencil for \( u^{n+1}_{i,j=0} \) to obtain a modified stencil on the boundary.
Pattern: use interior stencil also on the bundary, but replace \( j-1 \) by \( j+1 \)
Alternative: use ghost cells and ghost values
$$
\begin{align}
u_t &= c^2(u_{xx} + u_{yy}) + f(x,y,t),\quad &(x,y)\in \Omega,\ t\in (0,T]\\
u(x,y,0) &= I(x,y),\quad &(x,y)\in\Omega\\
u_t(x,y,0) &= V(x,y),\quad &(x,y)\in\Omega\\
u &= 0,\quad &(x,y)\in\partial\Omega,\ t\in (0,T]
\end{align}
$$
\( \Omega = [0,L_x]\times [0,L_y] \)
Discretization:
$$ [D_t D_t u = c^2(D_xD_x u + D_yD_y u) + f]^n_{i,j},
$$
Program: wave2D_u0.py
def solver(I, V, f, c, Lx, Ly, Nx, Ny, dt, T,
user_action=None, version='scalar'):
Mesh:
x = linspace(0, Lx, Nx+1) # mesh points in x dir
y = linspace(0, Ly, Ny+1) # mesh points in y dir
dx = x[1] - x[0]
dy = y[1] - y[0]
Nt = int(round(T/float(dt)))
t = linspace(0, N*dt, N+1) # mesh points in time
Cx2 = (c*dt/dx)**2; Cy2 = (c*dt/dy)**2 # help variables
dt2 = dt**2
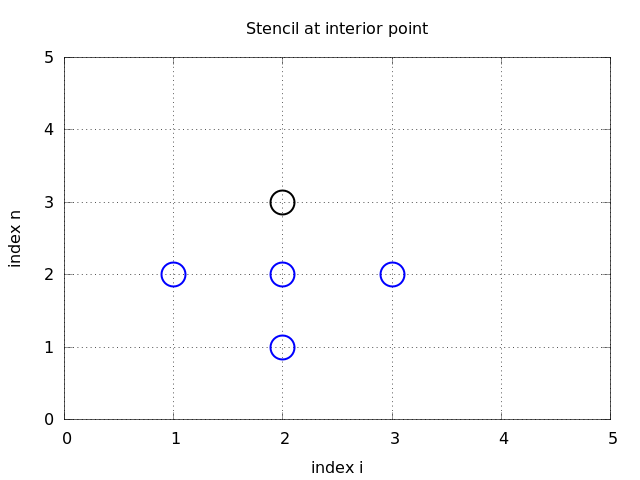
Store \( u^{n+1}_{i,j} \), \( u^{n}_{i,j} \), and \( u^{n-1}_{i,j} \) in three two-dimensional arrays:
u = zeros((Nx+1,Ny+1)) # solution array
u_1 = zeros((Nx+1,Ny+1)) # solution at t-dt
u_2 = zeros((Nx+1,Ny+1)) # solution at t-2*dt
\( u^{n+1}_{i,j} \) corresponds to u[i,j], etc.
Ix = range(0, u.shape[0])
Iy = range(0, u.shape[1])
It = range(0, t.shape[0])
for i in Ix:
for j in Iy:
u_1[i,j] = I(x[i], y[j])
if user_action is not None:
user_action(u_1, x, xv, y, yv, t, 0)
Arguments xv and yv: for vectorized computations
def advance_scalar(u, u_1, u_2, f, x, y, t, n, Cx2, Cy2, dt,
V=None, step1=False):
Ix = range(0, u.shape[0]); Iy = range(0, u.shape[1])
dt2 = dt**2
if step1:
Cx2 = 0.5*Cx2; Cy2 = 0.5*Cy2; dt2 = 0.5*dt2
D1 = 1; D2 = 0
else:
D1 = 2; D2 = 1
for i in Ix[1:-1]:
for j in Iy[1:-1]:
u_xx = u_1[i-1,j] - 2*u_1[i,j] + u_1[i+1,j]
u_yy = u_1[i,j-1] - 2*u_1[i,j] + u_1[i,j+1]
u[i,j] = D1*u_1[i,j] - D2*u_2[i,j] + \
Cx2*u_xx + Cy2*u_yy + dt2*f(x[i], y[j], t[n])
if step1:
u[i,j] += dt*V(x[i], y[j])
# Boundary condition u=0
j = Iy[0]
for i in Ix: u[i,j] = 0
j = Iy[-1]
for i in Ix: u[i,j] = 0
i = Ix[0]
for j in Iy: u[i,j] = 0
i = Ix[-1]
for j in Iy: u[i,j] = 0
return u
D1 and D2: allow advance_scalar to be used also for \( u^1_{i,j} \):
u = advance_scalar(u, u_1, u_2, f, x, y, t,
n, 0.5*Cx2, 0.5*Cy2, 0.5*dt2, D1=1, D2=0)
Mesh with \( 30\times 30 \) cells: vectorization reduces the CPU time by a factor of 70 (!).
Need special coordinate arrays xv and yv such that \( I(x,y) \)
and \( f(x,y,t) \) can be vectorized:
from numpy import newaxis
xv = x[:,newaxis]
yv = y[newaxis,:]
u_1[:,:] = I(xv, yv)
f_a[:,:] = f(xv, yv, t)
def advance_vectorized(u, u_1, u_2, f_a, Cx2, Cy2, dt,
V=None, step1=False):
dt2 = dt**2
if step1:
Cx2 = 0.5*Cx2; Cy2 = 0.5*Cy2; dt2 = 0.5*dt2
D1 = 1; D2 = 0
else:
D1 = 2; D2 = 1
u_xx = u_1[:-2,1:-1] - 2*u_1[1:-1,1:-1] + u_1[2:,1:-1]
u_yy = u_1[1:-1,:-2] - 2*u_1[1:-1,1:-1] + u_1[1:-1,2:]
u[1:-1,1:-1] = D1*u_1[1:-1,1:-1] - D2*u_2[1:-1,1:-1] + \
Cx2*u_xx + Cy2*u_yy + dt2*f_a[1:-1,1:-1]
if step1:
u[1:-1,1:-1] += dt*V[1:-1, 1:-1]
# Boundary condition u=0
j = 0
u[:,j] = 0
j = u.shape[1]-1
u[:,j] = 0
i = 0
u[i,:] = 0
i = u.shape[0]-1
u[i,:] = 0
return u
Manufactured solution:
$$
\begin{equation}
\uex(x,y,t) = x(L_x-x)y(L_y-y)(1+{\half}t)
\tag{43}
\end{equation}
$$
Requires \( f=2c^2(1+{\half}t)(y(L_y-y) + x(L_x-x)) \).
This \( \uex \) is ideal because it also solves the discrete equations!
$$
\begin{align*}
[D_xD_x \uex]^n_{i,j} &= [y(L_y-y)(1+{\half}t) D_xD_x x(L_x-x)]^n_{i,j}\\
&= y_j(L_y-y_j)(1+{\half}t_n)2
\end{align*}
$$
Pure Python code:
def advance_scalar(u, u_1, u_2, f, x, y, t,
n, Cx2, Cy2, dt2, D1=2, D2=1):
Ix = range(0, u.shape[0]); Iy = range(0, u.shape[1])
for i in Ix[1:-1]:
for j in Iy[1:-1]:
u_xx = u_1[i-1,j] - 2*u_1[i,j] + u_1[i+1,j]
u_yy = u_1[i,j-1] - 2*u_1[i,j] + u_1[i,j+1]
u[i,j] = D1*u_1[i,j] - D2*u_2[i,j] + \
Cx2*u_xx + Cy2*u_yy + dt2*f(x[i], y[j], t[n])
.pyx extension.function(a, b) \( \rightarrow \) cpdef function(int a, double b)v = 1.2 \( \rightarrow \) cdef double v = 1.2np.ndarray[np.float64_t, ndim=2, mode='c'] u
import numpy as np
cimport numpy as np
cimport cython
ctypedef np.float64_t DT # data type
@cython.boundscheck(False) # turn off array bounds check
@cython.wraparound(False) # turn off negative indices (u[-1,-1])
cpdef advance(
np.ndarray[DT, ndim=2, mode='c'] u,
np.ndarray[DT, ndim=2, mode='c'] u_1,
np.ndarray[DT, ndim=2, mode='c'] u_2,
np.ndarray[DT, ndim=2, mode='c'] f,
double Cx2, double Cy2, double dt2):
cdef int Nx, Ny, i, j
cdef double u_xx, u_yy
Nx = u.shape[0]-1
Ny = u.shape[1]-1
for i in xrange(1, Nx):
for j in xrange(1, Ny):
u_xx = u_1[i-1,j] - 2*u_1[i,j] + u_1[i+1,j]
u_yy = u_1[i,j-1] - 2*u_1[i,j] + u_1[i,j+1]
u[i,j] = 2*u_1[i,j] - u_2[i,j] + \
Cx2*u_xx + Cy2*u_yy + dt2*f[i,j]
Note: from now in we skip the code for setting boundary values
See how effective Cython can translate this code to C:
Terminal> cython -a wave2D_u0_loop_cy.pyx
Load wave2D_u0_loop_cy.html in a browser (white: pure C, yellow: still Python):
Can click on wave2D_u0_loop_cy.c to see the generated C code...
.so file) that can be
loaded as a standard Python modulesetup.py script to build the extension module
from distutils.core import setup
from distutils.extension import Extension
from Cython.Distutils import build_ext
cymodule = 'wave2D_u0_loop_cy'
setup(
name=cymodule
ext_modules=[Extension(cymodule, [cymodule + '.pyx'],)],
cmdclass={'build_ext': build_ext},
)
Terminal> python setup.py build_ext --inplace
import wave2D_u0_loop_cy
advance = wave2D_u0_loop_cy.advance
...
for n in It[1:-1: # time loop
f_a[:,:] = f(xv, yv, t[n]) # precompute, size as u
u = advance(u, u_1, u_2, f_a, x, y, t, Cx2, Cy2, dt2)
Efficiency:
numpy: 5.5
numpy: 6
advance function in pure Fortranf2py to generate C code for calling Fortran from Python
subroutine advance(u, u_1, u_2, f, Cx2, Cy2, dt2, Nx, Ny)
integer Nx, Ny
real*8 u(0:Nx,0:Ny), u_1(0:Nx,0:Ny), u_2(0:Nx,0:Ny)
real*8 f(0:Nx, 0:Ny), Cx2, Cy2, dt2
integer i, j
Cf2py intent(in, out) u
C Scheme at interior points
do j = 1, Ny-1
do i = 1, Nx-1
u(i,j) = 2*u_1(i,j) - u_2(i,j) +
& Cx2*(u_1(i-1,j) - 2*u_1(i,j) + u_1(i+1,j)) +
& Cy2*(u_1(i,j-1) - 2*u_1(i,j) + u_1(i,j+1)) +
& dt2*f(i,j)
end do
end do
Note: Cf2py comment declares u as input argument and return value
back to Python
Terminal> f2py -m wave2D_u0_loop_f77 -h wave2D_u0_loop_f77.pyf \
--overwrite-signature wave2D_u0_loop_f77.f
Terminal> f2py -c wave2D_u0_loop_f77.pyf --build-dir build_f77 \
-DF2PY_REPORT_ON_ARRAY_COPY=1 wave2D_u0_loop_f77.f
f2py changes the argument list (!)
>>> import wave2D_u0_loop_f77
>>> print wave2D_u0_loop_f77.__doc__
This module 'wave2D_u0_loop_f77' is auto-generated with f2py....
Functions:
u = advance(u,u_1,u_2,f,cx2,cy2,dt2,
nx=(shape(u,0)-1),ny=(shape(u,1)-1))
f2py!
f2py takes a copy of a numpy (C) array and transposes it
when calling Fortrannumpy arrays with Fortran storage
order = 'Fortran' if version == 'f77' else 'C'
u = zeros((Nx+1,Ny+1), order=order)
u_1 = zeros((Nx+1,Ny+1), order=order)
u_2 = zeros((Nx+1,Ny+1), order=order)
Option -DF2PY_REPORT_ON_ARRAY_COPY=1 makes f2py write out
array copying:
Terminal> f2py -c wave2D_u0_loop_f77.pyf --build-dir build_f77 \
-DF2PY_REPORT_ON_ARRAY_COPY=1 wave2D_u0_loop_f77.f
numpy code
advance function in pure C
numpy arrays transferred to C are one-dimensional in C[i,j] indices to single indices
/* Translate (i,j) index to single array index */
#define idx(i,j) (i)*(Ny+1) + j
void advance(double* u, double* u_1, double* u_2, double* f,
double Cx2, double Cy2, double dt2,
int Nx, int Ny)
{
int i, j;
/* Scheme at interior points */
for (i=1; i<=Nx-1; i++) {
for (j=1; j<=Ny-1; j++) {
u[idx(i,j)] = 2*u_1[idx(i,j)] - u_2[idx(i,j)] +
Cx2*(u_1[idx(i-1,j)] - 2*u_1[idx(i,j)] + u_1[idx(i+1,j)]) +
Cy2*(u_1[idx(i,j-1)] - 2*u_1[idx(i,j)] + u_1[idx(i,j+1)]) +
dt2*f[idx(i,j)];
}
}
}
}
import numpy as np
cimport numpy as np
cimport cython
cdef extern from "wave2D_u0_loop_c.h":
void advance(double* u, double* u_1, double* u_2, double* f,
double Cx2, double Cy2, double dt2,
int Nx, int Ny)
@cython.boundscheck(False)
@cython.wraparound(False)
def advance_cwrap(
np.ndarray[double, ndim=2, mode='c'] u,
np.ndarray[double, ndim=2, mode='c'] u_1,
np.ndarray[double, ndim=2, mode='c'] u_2,
np.ndarray[double, ndim=2, mode='c'] f,
double Cx2, double Cy2, double dt2):
advance(&u[0,0], &u_1[0,0], &u_2[0,0], &f[0,0],
Cx2, Cy2, dt2,
u.shape[0]-1, u.shape[1]-1)
return u
Compile and link the extension module with a setup.py file:
from distutils.core import setup
from distutils.extension import Extension
from Cython.Distutils import build_ext
sources = ['wave2D_u0_loop_c.c', 'wave2D_u0_loop_c_cy.pyx']
module = 'wave2D_u0_loop_c_cy'
setup(
name=module,
ext_modules=[Extension(module, sources,
libraries=[], # C libs to link with
)],
cmdclass={'build_ext': build_ext},
)
Terminal> python setup.py build_ext --inplace
In Python:
import wave2D_u0_loop_c_cy
advance = wave2D_u0_loop_c_cy.advance_cwrap
...
f_a[:,:] = f(xv, yv, t[n])
u = advance(u, u_1, u_2, f_a, Cx2, Cy2, dt2)
advance function in pure Cf2py to generate C code for calling C from Python
advance as beforeadvance functionf2py generate the Fortran 90 module
Fortran 77 signature (note intent(c)):
subroutine advance(u, u_1, u_2, f, Cx2, Cy2, dt2, Nx, Ny)
Cf2py intent(c) advance
integer Nx, Ny, N
real*8 u(0:Nx,0:Ny), u_1(0:Nx,0:Ny), u_2(0:Nx,0:Ny)
real*8 f(0:Nx, 0:Ny), Cx2, Cy2, dt2
Cf2py intent(in, out) u
Cf2py intent(c) u, u_1, u_2, f, Cx2, Cy2, dt2, Nx, Ny
return
end
Generate Fortran 90 module (wave2D_u0_loop_c_f2py.pyf):
Terminal> f2py -m wave2D_u0_loop_c_f2py \
-h wave2D_u0_loop_c_f2py.pyf --overwrite-signature \
wave2D_u0_loop_c_f2py_signature.f
The compile and build step must list the C files:
Terminal> f2py -c wave2D_u0_loop_c_f2py.pyf \
--build-dir tmp_build_c \
-DF2PY_REPORT_ON_ARRAY_COPY=1 wave2D_u0_loop_c.c
numpy C arrays to C++ array classesf2pynumpy arrays to C API and let C translate numpy arrays
into C++ array classes
$$
\begin{equation*} \frac{\partial^2 u}{\partial t^2} =
c^2 \frac{\partial^2 u}{\partial x^2}
\end{equation*}
$$
Solutions:
$$
\begin{equation}
u(x,t) = g_R(x-ct) + g_L(x+ct),
\tag{44}
\end{equation}
$$
If \( u(x,0)=I(x) \) and \( u_t(x,0)=0 \):
$$
\begin{equation}
u(x,t) = \half I(x-ct) + \half I(x+ct)
\tag{45}
\end{equation}
$$
Two waves: one traveling to the right and one to the left
A wave propagates perfectly (\( C=1 \)) and hits a medium with 1/4 of the wave velocity. A part of the wave is reflected and the rest is transmitted.
We have just changed the initial condition...
Build \( I(x) \) of wave components \( e^{ikx} = \cos kx + i\sin kx \):
$$
\begin{equation} I(x) \approx \sum_{k\in K} b_k e^{ikx}
\tag{46}
\end{equation}
$$
Since \( u(x,t)=\half I(x-ct) + \half I(x+ct) \):
$$
\begin{equation}
u(x,t) = \half \sum_{k\in K} b_k e^{ik(x - ct)}
+ \half \sum_{k\in K} b_k e^{ik(x + ct)}
\tag{47}
\end{equation}
$$
Our interest: one component \( e^{i(kx -\omega t)} \), \( \omega = kc \)
A similar discrete \( u^n_q = e^{i(kx_q - \tilde\omega t_n)} \) solves
$$
\begin{equation}
[D_tD_t u = c^2 D_xD_x u]^n_q
\tag{48}
\end{equation}
$$
Note: different frequency \( \tilde\omega\neq\omega \)
$$
\begin{equation*}
[D_tD_t e^{i\omega t}]^n = -\frac{4}{\Delta t^2}\sin^2\left(
\frac{\omega\Delta t}{2}\right)e^{i\omega n\Delta t}
\end{equation*}
$$
By \( \omega\rightarrow k \), \( t\rightarrow x \), \( n\rightarrow q \)) it follows that
$$
\begin{equation*}
[D_xD_x e^{ikx}]_q = -\frac{4}{\Delta x^2}\sin^2\left(
\frac{k\Delta x}{2}\right)e^{ikq\Delta x}
\end{equation*}
$$
Inserting a basic wave component \( u=e^{i(kx_q-\tilde\omega t_n)} \) in the scheme (48) requires computation of
$$
\begin{align}
\lbrack D_tD_t e^{ikx}e^{-i\tilde\omega t}\rbrack^n_q &= \lbrack D_tD_t e^{-i\tilde\omega t}\rbrack^ne^{ikq\Delta x}\nonumber\\ &= -\frac{4}{\Delta t^2}\sin^2\left(
\frac{\tilde\omega\Delta t}{2}\right)e^{-i\tilde\omega n\Delta t}e^{ikq\Delta x}\\
\lbrack D_xD_x e^{ikx}e^{-i\tilde\omega t}\rbrack^n_q &= \lbrack D_xD_x e^{ikx}\rbrack_q e^{-i\tilde\omega n\Delta t}\nonumber\\ &= -\frac{4}{\Delta x^2}\sin^2\left(
\frac{k\Delta x}{2}\right)e^{ikq\Delta x}e^{-i\tilde\omega n\Delta t} \end{align}
$$
The complete scheme,
$$
\begin{equation*}
\lbrack D_tD_t e^{ikx}e^{-i\tilde\omega t} = c^2D_xD_x e^{ikx}e^{-i\tilde\omega t}\rbrack^n_q
\end{equation*}
$$
leads to an equation for \( \tilde\omega \):
$$
\begin{equation}
\sin^2\left(\frac{\tilde\omega\Delta t}{2}\right)
= C^2\sin^2\left(\frac{k\Delta x}{2}\right),
\tag{49}
\end{equation}
$$
where \( C = \frac{c\Delta t}{\Delta x} \) is the Courant number
Taking the square root of (49):
$$
\begin{equation}
\sin\left(\frac{\tilde\omega\Delta t}{2}\right)
= C\sin\left(\frac{k\Delta x}{2}\right),
\tag{50}
\end{equation}
$$
Stability criterion
$$
\begin{equation}
C = \frac{c\Delta t}{\Delta x} \leq 1
\tag{51}
\end{equation}
$$
Assume \( C>1 \). Then
$$
\underbrace{\sin\left(\frac{\tilde\omega\Delta t}{2}\right)}{>1} = C\sin\left(\frac{k\Delta x}{2}\right)
$$
$$
\begin{equation}
\tilde\omega = \frac{2}{\Delta t}
\sin^{-1}\left( C\sin\left(\frac{k\Delta x}{2}\right)\right)
\tag{52}
\end{equation}
$$
$$
\begin{equation*}
r(C, p) = \frac{\tilde c}{c} = \frac{1}{Cp}{\sin}^{-1}\left(C\sin p\right),
\quad C\in (0,1],\ p\in (0,\pi/2]
\end{equation*}
$$
def r(C, p):
return 2/(C*p)*asin(C*sin(p))
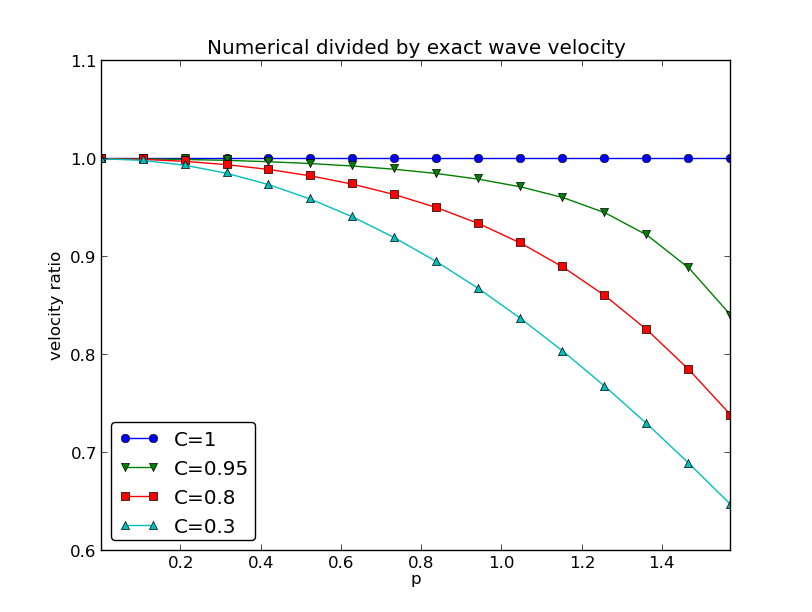
Note: the shortest waves have the largest error, and short waves move too slowly.
For small \( p \), Taylor expand \( \tilde\omega \) as polynomial in \( p \):
>>> C, p = symbols('C p')
>>> rs = r(C, p).series(p, 0, 7)
>>> print rs
1 - p**2/6 + p**4/120 - p**6/5040 + C**2*p**2/6 -
C**2*p**4/12 + 13*C**2*p**6/720 + 3*C**4*p**4/40 -
C**4*p**6/16 + 5*C**6*p**6/112 + O(p**7)
>>> # Factorize each term and drop the remainder O(...) term
>>> rs_factored = [factor(term) for term in rs.lseries(p)]
>>> rs_factored = sum(rs_factored)
>>> print rs_factored
p**6*(C - 1)*(C + 1)*(225*C**4 - 90*C**2 + 1)/5040 +
p**4*(C - 1)*(C + 1)*(3*C - 1)*(3*C + 1)/120 +
p**2*(C - 1)*(C + 1)/6 + 1
Leading error term is \( \frac{1}{6}(C^2-1)p^2 \) or
$$
\begin{equation}
\frac{1}{6}\left(\frac{k\Delta x}{2}\right)^2(C^2-1)
= \frac{k^2}{24}\left( c^2\Delta t^2 - \Delta x^2\right) =
\Oof{\Delta t^2, \Delta x^2}
\end{equation}
$$
Smooth wave, few short waves (large \( k \)) in \( I(x) \):
Not so smooth wave, significant short waves (large \( k \)) in \( I(x) \):
$$ u(x,y,t) = g(k_xx + k_yy - \omega t) $$
is a typically solution of
$$ u_{tt} = c^2(u_{xx} + u_{yy}) $$
Can build solutions by adding complex Fourier components of the form
$$
e^{i(k_xx + k_yy - \omega t)}
$$
$$
\begin{equation}
\lbrack D_tD_t u = c^2(D_xD_x u + D_yD_y u)\rbrack^n_{q,r}
\tag{53}
\end{equation}
$$
This equation admits a Fourier component
$$
\begin{equation}
u^n_{q,r} = e^{i(k_x q\Delta x + k_y r\Delta y
- \tilde\omega n\Delta t)}
\tag{54}
\end{equation}
$$
Inserting the expression and using formulas from the 1D analysis:
$$
\begin{equation}
\sin^2\left(\frac{\tilde\omega\Delta t}{2}\right)
= C_x^2\sin^2 p_x
+ C_y^2\sin^2 p_y, \end{equation}
$$
where
$$ C_x = \frac{c^2\Delta t^2}{\Delta x^2},\quad
C_y = \frac{c^2\Delta t^2}{\Delta y^2}, \quad
p_x = \frac{k_x\Delta x}{2},\quad
p_y = \frac{k_y\Delta y}{2}
$$
Rreal-valued \( \tilde\omega \) requires
$$
\begin{equation}
C_x^2 + C_y^2 \leq 1
\tag{55}
\end{equation}
$$
or
$$
\begin{equation}
\Delta t \leq \frac{1}{c} \left( \frac{1}{\Delta x^2} +
\frac{1}{\Delta y^2}\right)^{-\halfi}
\tag{56}
\end{equation}
$$
$$
\begin{equation}
\Delta t \leq \frac{1}{c}\left( \frac{1}{\Delta x^2} +
\frac{1}{\Delta y^2} + \frac{1}{\Delta z^2}\right)^{-\halfi}
\end{equation}
$$
For \( c^2=c^2(\xpoint) \) we must use the worst-case value \( \bar c = \sqrt{\max_{\xpoint\in\Omega} c^2(\xpoint)} \) and a safety factor \( \beta\leq 1 \):
$$
\begin{equation}
\Delta t \leq \beta \frac{1}{\bar c}
\left( \frac{1}{\Delta x^2} +
\frac{1}{\Delta y^2} + \frac{1}{\Delta z^2}\right)^{-\halfi}
\end{equation}
$$
$$
\tilde\omega = \frac{2}{\Delta t}\sin^{-1}\left(
\left( C_x^2\sin^2 p_x + C_y^2\sin^ p_y\right)^\half\right)
$$
For visualization, introduce \( \theta \):
$$ k_x = k\sin\theta,\quad k_y=k\cos\theta,
\quad p_x=\half kh\cos\theta,\quad p_y=\half kh\sin\theta$$
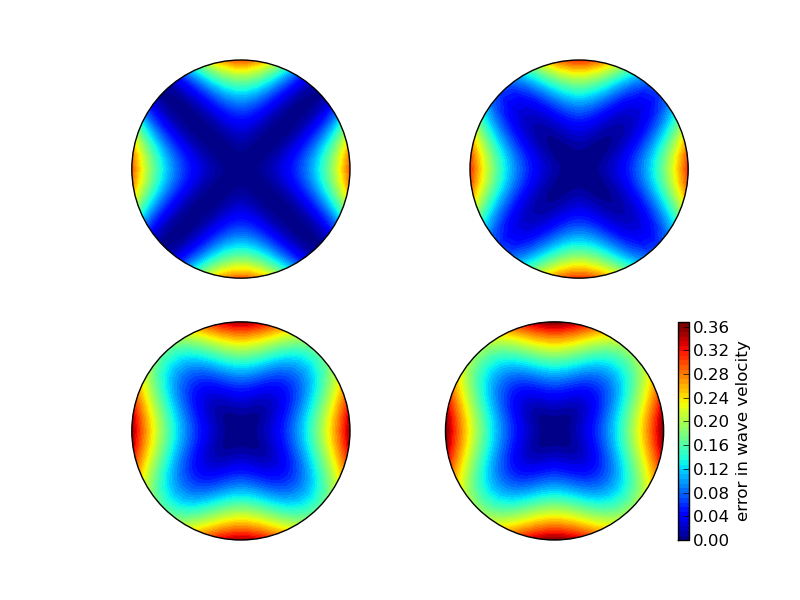
Also: \( \Delta x=\Delta y=h \). Then \( C_x=C_y=c\Delta t/h\equiv C \).
Now \( \tilde\omega \) depends on
$$ \frac{\tilde c}{c} = \frac{1}{Ckh}
\sin^{-1}\left(C\left(\sin^2 ({\half}kh\cos\theta)
+ \sin^2({\half}kh\sin\theta) \right)^\half\right)
$$
Can make color contour plots of \( 1-\tilde c/c \) in polar coordinates with \( \theta \) as the angular coordinate and \( kh \) as the radial coordinate.