Study Guide: Intro to Computing with Finite Difference Methods
Dec 14, 2013
INF5620 in a nutshell
- Numerical methods for partial differential equations (PDEs)
- How to we solve a PDE in practice and produce numbers?
- How to we trust the answer?
- Approach: simplify, understand, generalize
The new official six-point course description
After having completed INF5620 you
- can derive methods and implement them to solve frequently arising partial differential equations (PDEs) from physics and mechanics.
- have a good understanding of finite difference and finite element methods and how they are applied in linear and nonlinear PDE problems.
- can identify numerical artifacts and perform mathematical analysis to understand and cure non-physical effects.
- can apply sophisticated programming techniques in Python, combined with Cython, C, C++, and Fortran code, to create modern, flexible simulation programs.
- can construct verification tests and automate them.
- have experience with project hosting sites (Bitbucket, GitHub), version control systems (Git), report writing (LaTeX), and Python scripting for performing reproducible computational science.
More specific description of the contents; part 1
- Finite difference methods
- ODEs
- the wave equation \( u_{tt}=u_{xx} \) in 1D, 2D, 3D
- the diffusion equation \( u_t=u_{xx} \) in 1D, 2D, 3D
- write your own software from scratch
- understand how the methods work and why they fail
- Finite element methods for
- stationary diffusion equations \( u_{xx}=f \) in 1D
- time-dependent diffusion and wave equations in 1D
- PDEs in 2D and 3D by use of the FEniCS software
- perform hand-calculations, write your own software (1D)
- understand how the methods work and why they fail
More specific description of the contents; part 2
- Nonlinear PDEs
- Newton and Picard iteration methods, finite differences and elements
- More advanced PDEs for fluid flow and elasticity
- Parallel computing
Philosophy: simplify, understand, generalize
- Start with simplified ODE/PDE problems
- Learn to reason about the discretization
- Learn to implement, verify, and experiment
- Understand the method, program, and results
- Generalize the problem, method, and program
This is the power of applied mathematics!
The exam
- Oral exam
- 6 problems (topics) are announced two weeks before the exam
- Work out a 20 min presentations (talks) for each problem
- At the exam: throw a die to pick your problem to be presented
- Aids: plots, computer programs
- Why? Very effective way of learning
- Sure? Excellent results over 15 years
- When? Late december
Required software
- Our software platform: Python (sometimes combined with Cython, Fortran, C, C++)
- Important Python packages:
numpy,scipy,matplotlib,sympy,fenics,scitools, ... - Suggested installation: Run Ubuntu in a virtual machine
- Alternative: run a (course-specific) Vagrant machine
Assumed/ideal background
- INF1100: Python programming, solution of ODEs
- Some experience with finite difference methods
- Some analytical and numerical knowledge of PDEs
- Much experience with calculus and linear algebra
- Much experience with programming of mathematical problems
- Experience with mathematical modeling with PDEs (from physics, mechanics, geophysics, or ...)
Start-up example for the course
What if you don't have this ideal background?
- Students come to this course with very different backgrounds
- First task: summarize assumed background knowledge by going through a simple example
- Also in this example:
- Some fundamental material on software implementation and software testing
- Material on analyzing numerical methods to understand why they can fail
- Applications to real-world problems
Start-up example
$$ u'=-au,\quad u(0)=I,\ t\in (0,T],$$
where \( a>0 \) is a constant.
Everything we do is motivated by what we need as building blocks for solving PDEs!
What to learn in the start-up example; standard topics
- How to think when constructing finite difference methods, with special focus on the Forward Euler, Backward Euler, and Crank-Nicolson (midpoint) schemes
- How to formulate a computational algorithm and translate it into Python code
- How to make curve plots of the solutions
- How to compute numerical errors
- How to compute convergence rates
What to learn in the start-up example; programming topics
- How to verify an implementation and automate verification through nose tests in Python
- How to structure code in terms of functions, classes, and modules
- How to work with Python concepts such as arrays, lists, dictionaries, lambda functions, functions in functions (closures), doctests, unit tests, command-line interfaces, graphical user interfaces
- How to perform array computing and understand the difference from scalar computing
- How to conduct and automate large-scale numerical experiments
- How to generate scientific reports
What to learn in the start-up example; mathematical analysis
- How to uncover numerical artifacts in the computed solution
- How to analyze the numerical schemes mathematically to understand why artifacts occur
- How to derive mathematical expressions for various measures of
the error in numerical methods, frequently by using the
sympysoftware for symbolic computation - Introduce concepts such as finite difference operators, mesh (grid), mesh functions, stability, truncation error, consistency, and convergence
What to learn in the start-up example; generalizations
- Generalize the example to \( u'(t)=-a(t)u(t) + b(t) \)
- Present additional methods for the general nonlinear ODE \( u'=f(u,t) \), which is either a scalar ODE or a system of ODEs
- How to access professional packages for solving ODEs
- How our model equations like \( u'=-au \) arises in a wide range of phenomena in physics, biology, and finance
Finite difference methods
- The finite difference method is the simplest method for solving differential equations
- Fast to learn, derive, and implement
- A very useful tool to know, even if you aim at using the finite element or the finite volume method
Topics in the first intro to the finite difference method
- How to derive a finite difference discretization of an ODE
- Key concepts: mesh, mesh function, finite difference approximations
- The Forward Euler, Backward Euler, and Crank-Nicolson methods
- Finite difference operator notation
- How to derive an algorithm and implement it in Python
- How to test the implementation
A basic model for exponential decay
The world's simplest (?) ODE:
$$
\begin{equation*}
u'(t) = -au(t),\quad u(0)=I,\ t\in (0,T]\tp
\end{equation*}
$$
Applications
- Growth and decay of populations (cells, animals, human)
- Growth and decay of a fortune
- Radioactive decay
- Cooling/heating of an object
- Pressure variation in the atmosphere
- Vertical motion of a body in water/air
- Time-discretization of diffusion PDEs by Fourier techniques
See the text for details.
Continuous problem
$$
\begin{equation}
u' = -au,\ t\in (0,T], \quad u(0)=I\tp \tag{1}
\end{equation}
$$
Solution of the continuous problem ("continuous solution"):
$$
\begin{equation*} u(t) = Ie^{-at}\tp\end{equation*}
$$
(special case that we can derive a formula for the discrete solution)
Discrete problem
\( u^n\approx u(t_n) \) means that \( u \) is found at discrete time points \( t_1,t_2,t_3,\ldots \)
Typical computational formula:
$$
\begin{equation*} u^{n+1} = Au^n\tp\end{equation*}
$$
The constant \( A \) depends on the type of finite difference method.
Solution of the discrete problem ("discrete solution"):
$$
\begin{equation*} u^{n+1} = IA^n\tp\end{equation*}
$$
(special case that we can derive a formula for the discrete solution)
The steps in the finite difference method
Solving a differential equation by a finite difference method consists of four steps:
- discretizing the domain,
- fulfilling the equation at discrete time points,
- replacing derivatives by finite differences,
- formulating a recursive algorithm.
Step 1: Discretizing the domain
The time domain \( [0,T] \) is represented by a mesh: a finite number of \( N_t+1 \) points
$$0 = t_0 < t_1 < t_2 < \cdots < t_{N_t-1} < t_{N_t} = T\tp$$
- We seek the solution \( u \) at the mesh points: \( u(t_n) \), \( n=1,2,\ldots,N_t \).
- Note: \( u^0 \) is known as \( I \).
- Notational short-form for the numerical approximation to \( u(t_n) \): \( u^n \)
- In the differential equation: \( u \) is the exact solution
- In the numerical method and implementation: \( u^n \) is the numerical approximation, \( \uex(t) \) is the exact solution
Step 1: Discretizing the domain
\( u^n \) is a mesh function, defined at the mesh points \( t_n \), \( n=0,\ldots,N_t \) only.
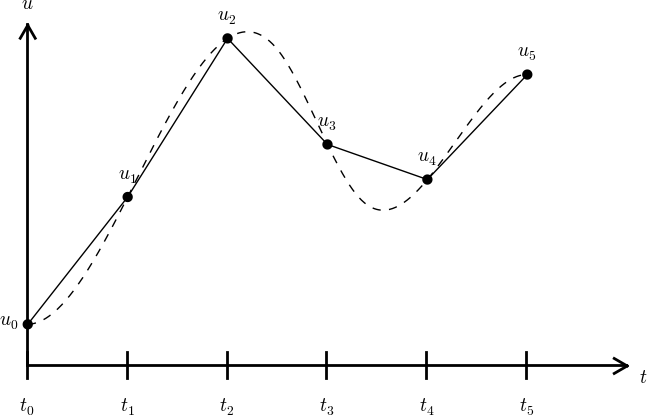
What about a mesh function between the mesh points?
Can extend the mesh function to yield values between mesh points by linear interpolation:
$$
\begin{equation}
u(t) \approx u^n + \frac{u^{n+1}-u^n}{t_{n+1}-t_n}(t - t_n)\tp
\end{equation}
$$

Step 2: Fulfilling the equation at discrete time points
- The ODE holds for all \( t\in (0,T] \) (infinite no of points)
- Idea: let the ODE be valid at the mesh points only (finite no of points)
$$
\begin{equation}
u'(t_n) = -au(t_n),\quad n=1,\ldots,N_t\tp
\tag{2}
\end{equation}
$$
Step 3: Replacing derivatives by finite differences
Now it is time for the finite difference approximations of derivatives:
$$
\begin{equation}
u'(t_n) \approx \frac{u^{n+1}-u^{n}}{t_{n+1}-t_n}\tp
\tag{3}
\end{equation}
$$

Step 3: Replacing derivatives by finite differences
Inserting the finite difference approximation in
$$ u'(t_n) = -au(t_n),$$
gives
$$
\begin{equation}
\frac{u^{n+1}-u^{n}}{t_{n+1}-t_n} = -au^{n},\quad n=0,1,\ldots,N_t-1\tp
\tag{4}
\end{equation}
$$
This is the
- discrete equation
- discrete problem
- finite difference method
- finite difference scheme
Step 4: Formulating a recursive algorithm
- How can we actually compute the \( u^n \) values?
- Fundamental structure:
- given \( u^0=I \)
- compute \( u^1 \) from \( u^0 \)
- compute \( u^2 \) from \( u^1 \)
- compute \( u^3 \) from \( u^2 \) (and so forth)
- In general: we have \( u^n \) and seek \( u^{n+1} \)
$$
\begin{equation}
u^{n+1} = u^n - a(t_{n+1} -t_n)u^n\tp
\tag{5}
\end{equation}
$$
Let us apply the scheme
Assume constant time spacing: \( \Delta t = t_{n+1}-t_n=\mbox{const} \)
$$
\begin{align*}
u_0 &= I,\\
u_1 & = u^0 - a\Delta t u^0 = I(1-a\Delta t),\\
u_2 & = I(1-a\Delta t)^2,\\
u^3 &= I(1-a\Delta t)^3,\\
&\vdots\\
u^{N_t} &= I(1-a\Delta t)^{N_t}\tp
\end{align*}
$$
Ooops - we can find the numerical solution by hand (in this simple example)! No need for a computer (yet)...
A backward difference
Here is another finite difference approximation to the derivative (backward difference):
$$
\begin{equation}
u'(t_n) \approx \frac{u^{n}-u^{n-1}}{t_{n}-t_{n-1}}\tp
\tag{6}
\end{equation}
$$

The Backward Euler scheme
Inserting the finite difference approximation in \( u'(t_n)=-au(t_n) \) yields the Backward Euler (BE) scheme:
$$
\begin{equation}
\frac{u^{n}-u^{n-1}}{t_{n}-t_{n-1}} = -a u^n\tp
\tag{7}
\end{equation}
$$
Solve with respect to the unknown \( u^{n+1} \):
$$
\begin{equation}
u^{n+1} = \frac{1}{1+ a(t_{n+1}-t_n)} u^n\tp
\tag{8}
\end{equation}
$$
A centered difference
Centered differences are better approximations than forward or backward differences.
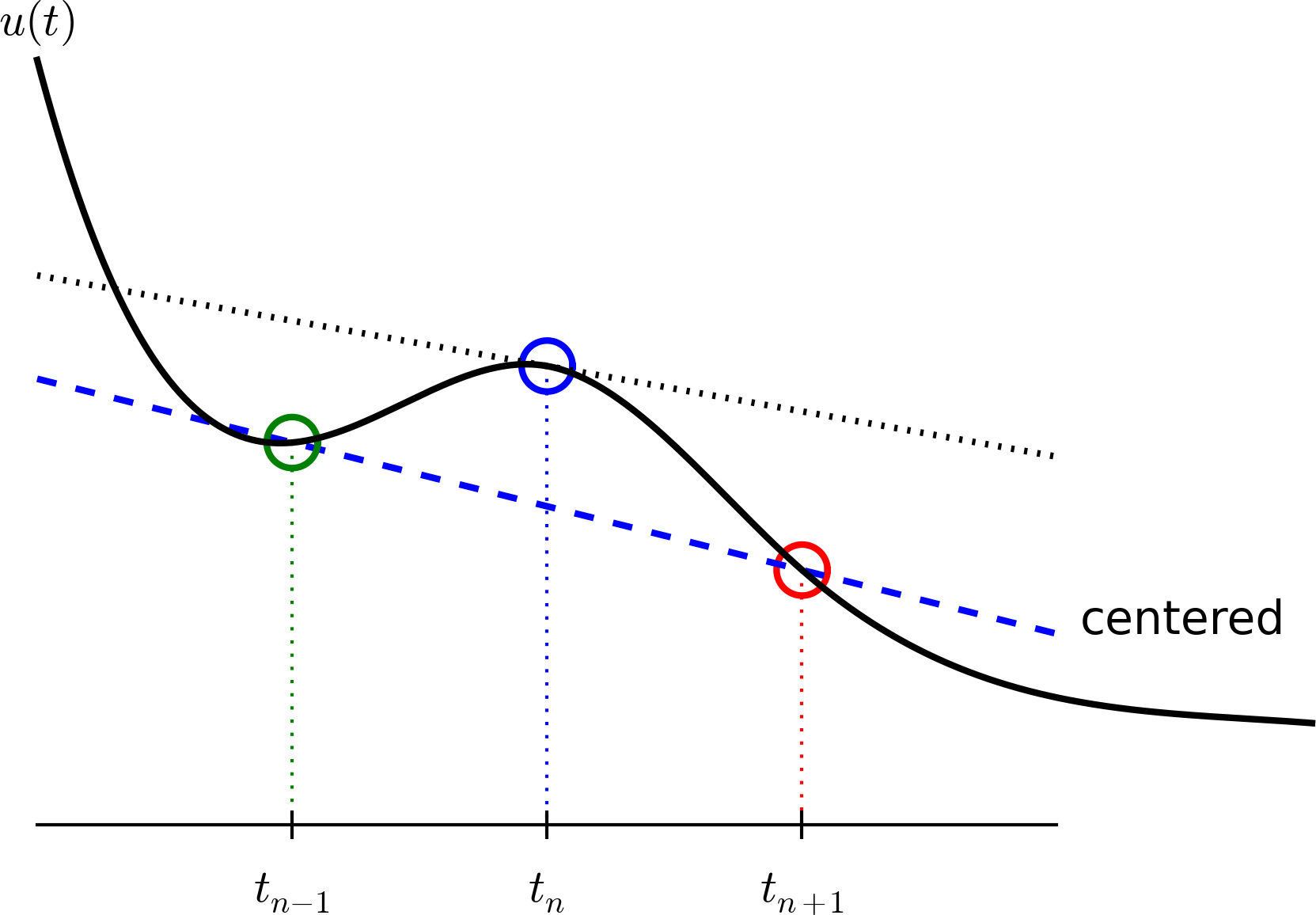
The Crank-Nicolson scheme; part 1
Idea 1: let the ODE hold at \( t_{n+1/2} \)
$$ u'(t_{n+1/2} = -au(t_{n+1/2})\tp$$
Idea 2: approximate \( u'(t_{n+1/2} \) by a centered difference
$$
\begin{equation}
u'(t_{n+\half}) \approx \frac{u^{n+1}-u^n}{t_{n+1}-t_n}\tp
\tag{9}
\end{equation}
$$
Problem: \( u(t_{n+1/2}) \) is not defined, only \( u^n=u(t_n) \) and \( u^{n+1}=u(t_{n+1}) \)
Solution:
$$ u(t_{n+1/2}) \approx \half(u^n + u^{n+1}) $$
The Crank-Nicolson scheme; part 2
Result:
$$
\begin{equation}
\frac{u^{n+1}-u^n}{t_{n+1}-t_n} = -a\half (u^n + u^{n+1})\tp
\tag{10}
\end{equation}
$$
Solve wrt to \( u^{n+1} \):
$$
\begin{equation}
u^{n+1} = \frac{1-\half a(t_{n+1}-t_n)}{1 + \half a(t_{n+1}-t_n)}u^n\tp
\tag{11}
\end{equation}
$$
This is a Crank-Nicolson (CN) scheme or a midpoint or centered scheme.
The unifying \( \theta \)-rule
The Forward Euler, Backward Euler, and Crank-Nicolson schemes can be formulated as one scheme with a varying parameter \( \theta \):
$$
\begin{equation}
\frac{u^{n+1}-u^{n}}{t_{n+1}-t_n} = -a (\theta u^{n+1} + (1-\theta) u^{n})
\tag{12}
\tp
\end{equation}
$$
- \( \theta =0 \): Forward Euler
- \( \theta =1 \): Backward Euler
- \( \theta =1/2 \): Crank-Nicolson
- We may alternatively choose any \( \theta\in [0,1] \).
\( u^n \) is known, solve for \( u^{n+1} \):
$$
\begin{equation}
u^{n+1} = \frac{1 - (1-\theta) a(t_{n+1}-t_n)}{1 + \theta a(t_{n+1}-t_n)}\tp
\tag{13}
\end{equation}
$$
Constant time step
Very common assumption (not important, but exclusively used for simplicity hereafter): constant time step \( t_{n+1}-t_n\equiv\Delta t \)
$$
\begin{align}
u^{n+1} &= (1 - a\Delta t )u^n \quad (\hbox{FE})
\tag{14}\\
u^{n+1} &= \frac{1}{1+ a\Delta t} u^n \quad (\hbox{BE})
\tag{15}\\
u^{n+1} &= \frac{1-\half a\Delta t}{1 + \half a\Delta t} u^n \quad (\hbox{CN})
\tag{16}\\
u^{n+1} &= \frac{1 - (1-\theta) a\Delta t}{1 + \theta a\Delta t}u^n \quad (\theta-\hbox{rule})
\tag{17}
\end{align}
$$
Test the understanding!
Derive Forward Euler, Backward Euler, and Crank-Nicolson schemes for Newton's law of cooling:
$$ T' = -k(T-T_s),\quad T(0)=T_0,\ t\in (0,t_{\mbox{end}}]\tp$$
Physical quantities:
- \( T(t) \): temperature of an object at time \( t \)
- \( k \): parameter expressing heat loss to the surroundings
- \( T_s \): temperature of the surroundings
- \( T_0 \): initial temperature
Compact operator notation for finite differences
- Finite difference formulas can be tedious to write and read/understand
- Handy tool: finite difference operator notation
- Advantage: communicates the nature of the difference in a compact way
$$
\begin{equation}
[D_t^- u = -au]^n \tp
\end{equation}
$$
Compact operator notation for difference operators
Forward difference:
$$
\begin{equation}
[D_t^+u]^n = \frac{u^{n+1} - u^{n}}{\Delta t}
\approx \frac{d}{dt} u(t_n) \tag{18}
\tp
\end{equation}
$$
Centered difference:
$$
\begin{equation}
[D_tu]^n = \frac{u^{n+\half} - u^{n-\half}}{\Delta t}
\approx \frac{d}{dt} u(t_n), \tag{19}
\end{equation}
$$
Backward difference:
$$
\begin{equation}
[D_t^-u]^n = \frac{u^{n} - u^{n-1}}{\Delta t}
\approx \frac{d}{dt} u(t_n) \tag{20}
\end{equation}
$$
The Backward Euler scheme with operator notation
$$
\begin{equation*}
[D_t^-u]^n = -au^n \tp
\end{equation*}
$$
Common to put the whole equation inside square brackets:
$$
\begin{equation}
[D_t^- u = -au]^n \tp
\end{equation}
$$
The Forward Euler scheme with operator notation
$$
\begin{equation}
[D_t^+ u = -au]^n\tp
\end{equation}
$$
The Crank-Nicolson scheme with operator notation
Introduce an averaging operator:
$$
\begin{equation}
[\overline{u}^{t}]^n = \half (u^{n-\half} + u^{n+\half} )
\approx u(t_n) \tag{21}
\end{equation}
$$
The Crank-Nicolson scheme can then be written as
$$
\begin{equation}
[D_t u = -a\overline{u}^t]^{n+\half}\tp
\tag{22}
\end{equation}
$$
Test: use the definitions and write out the above formula to see that it really is the Crank-Nicolson scheme!
Implementation
Model:
$$
u'(t) = -au(t),\quad t\in (0,T], \quad u(0)=I,
$$
Numerical method:
$$
u^{n+1} = \frac{1 - (1-\theta) a\Delta t}{1 + \theta a\Delta t}u^n,
$$
for \( \theta\in [0,1] \). Note
- \( \theta=0 \) gives Forward Euler
- \( \theta=1 \) gives Backward Euler
- \( \theta=1/2 \) gives Crank-Nicolson
Requirements of a program
- Compute the numerical solution \( u^n \), \( n=1,2,\ldots,N_t \)
- Display the numerical and exact solution \( \uex(t)=e^{-at} \)
- Bring evidence to a correct implementation (verification)
- Compare the numerical and the exact solution in a plot
- computes the error \( \uex (t_n) - u^n \)
- computes the convergence rate of the numerical scheme
- reads its input data from the command line
Tools to learn
- Basic Python programming
- Array computing with numpy
- Plotting with matplotlib.pyplot and scitools
- File writing and reading
- Making command-line user interface via
argparse.ArgumentParser - Making graphical user interfaces via Parampool
Why implement in Python?
- Python has a very clean, readable syntax (often known as "executable pseudo-code").
- Python code is very similar to MATLAB code (and MATLAB has a particularly widespread use for scientific computing).
- Python is a full-fledged, very powerful programming language.
- Python is similar to, but much simpler to work with and results in more reliable code than C++.
Why implement in Python?
- Python has a rich set of modules for scientific computing, and its popularity in scientific computing is rapidly growing.
- Python was made for being combined with compiled languages (C, C++, Fortran) to reuse existing numerical software and to reach high computational performance of new implementations.
- Python has extensive support for administrative task needed when doing large-scale computational investigations.
- Python has extensive support for graphics (visualization, user interfaces, web applications).
- FEniCS, a very powerful tool for solving PDEs by the finite element method, is most human-efficient to operate from Python.
Algorithm
- Store \( u^n \), \( n=0,1,\ldots,N_t \) in an array
u. - Algorithm:
- initialize \( u^0 \)
- for \( t=t_n \), \( n=1,2,\ldots,N_t \): compute \( u_n \) using the \( \theta \)-rule formula
Translation to Python function
from numpy import *
def solver(I, a, T, dt, theta):
"""Solve u'=-a*u, u(0)=I, for t in (0,T] with steps of dt."""
Nt = int(T/dt) # no of time intervals
T = Nt*dt # adjust T to fit time step dt
u = zeros(Nt+1) # array of u[n] values
t = linspace(0, T, Nt+1) # time mesh
u[0] = I # assign initial condition
for n in range(0, Nt): # n=0,1,...,Nt-1
u[n+1] = (1 - (1-theta)*a*dt)/(1 + theta*dt*a)*u[n]
return u, t
Note about the for loop: range(0, Nt, s) generates all integers
from 0 to Nt in steps of s (default 1), but not including Nt (!).
Sample call:
u, t = solver(I=1, a=2, T=8, dt=0.8, theta=1)
Integer division
Python applies integer division: 1/2 is 0, while 1./2 or 1.0/2 or
1/2. or 1/2.0 or 1.0/2.0 all give 0.5.
A safer solver function (dt = float(dt) - guarantee float):
from numpy import *
def solver(I, a, T, dt, theta):
"""Solve u'=-a*u, u(0)=I, for t in (0,T] with steps of dt."""
dt = float(dt) # avoid integer division
Nt = int(round(T/dt)) # no of time intervals
T = Nt*dt # adjust T to fit time step dt
u = zeros(Nt+1) # array of u[n] values
t = linspace(0, T, Nt+1) # time mesh
u[0] = I # assign initial condition
for n in range(0, Nt): # n=0,1,...,Nt-1
u[n+1] = (1 - (1-theta)*a*dt)/(1 + theta*dt*a)*u[n]
return u, t
Doc strings
- First string after the function heading
- Used for documenting the function
- Automatic documentation tools can make fancy manuals for you
- Can be used for automatic testing
def solver(I, a, T, dt, theta):
"""
Solve
u'(t) = -a*u(t),
with initial condition u(0)=I, for t in the time interval
(0,T]. The time interval is divided into time steps of
length dt.
theta=1 corresponds to the Backward Euler scheme, theta=0
to the Forward Euler scheme, and theta=0.5 to the Crank-
Nicolson method.
"""
...
Formatting of numbers
Can control formatting of reals and integers through the printf format:
print 't=%6.3f u=%g' % (t[i], u[i])
Or the alternative format string syntax:
print 't={t:6.3f} u={u:g}'.format(t=t[i], u=u[i])
Running the program
How to run the program decay_v1.py:
Terminal> python decay_v1.py
Can also run it as "normal" Unix programs: ./decay_v1.py if the
first line is
`#!/usr/bin/env python`
Then
Terminal> chmod a+rx decay_v1.py
Terminal> ./decay_v1.py
Verifying the implementation
- Verification = bring evidence that the program works
- Find suitable test problems
- Make function for each test problem
- Later: put the verification tests in a professional testing framework
Simplest method: run a few algorithmic steps by hand
Use a calculator (\( I=0.1 \), \( \theta=0.8 \), \( \Delta t =0.8 \)):
$$ A\equiv \frac{1 - (1-\theta) a\Delta t}{1 + \theta a \Delta t} = 0.298245614035$$
$$
\begin{align*}
u^1 &= AI=0.0298245614035,\\
u^2 &= Au^1= 0.00889504462912,\\
u^3 &=Au^2= 0.00265290804728
\end{align*}
$$
See the function verify_three_steps in decay_verf1.py.
Comparison with an exact discrete solution
Define
$$ A = \frac{1 - (1-\theta) a\Delta t}{1 + \theta a \Delta t}\tp $$
Repeated use of the \( \theta \)-rule:
$$
\begin{align*}
u^0 &= I,\\
u^1 &= Au^0 = AI,\\
u^n &= A^nu^{n-1} = A^nI \tp
\end{align*}
$$
Making a test based on an exact discrete solution
The exact discrete solution as
$$
\begin{equation}
u^n = IA^n
\tag{23}
\tp
\end{equation}
$$
Test if
$$ \max_n |u^n - \uex(t_n)| < \epsilon\sim 10^{-15}$$
Implementation in decay_verf2.py.
Test the understanding!
Make a program for solving Newton's law of cooling
$$ T' = -k(T-T_s),\quad T(0)=T_0,\ t\in (0,t_{\mbox{end}}]\tp$$
with the Forward Euler, Backward Euler, and Crank-Nicolson schemes
(or a \( \theta \) scheme). Verify the implementation.
Computing the numerical error as a mesh function
Task: compute the numerical error \( e^n = \uex(t_n) - u^n \)
Exact solution: \( \uex(t)=Ie^{-at} \), implemented as
def exact_solution(t, I, a):
return I*exp(-a*t)
Compute \( e^n \) by
u, t = solver(I, a, T, dt, theta) # Numerical solution
u_e = exact_solution(t, I, a)
e = u_e - u
-
exact_solution(t, I, a)works withtas array - Must have
expfromnumpy(notmath) -
e = u_e - u: array subtraction - Array arithmetics gives shorter and much faster code
Computing the norm of the error
- \( e^n \) is a mesh function
- Usually we want one number for the error
- Use a norm of \( e^n \)
Norms of a function \( f(t) \):
$$
\begin{align}
||f||_{L^2} &= \left( \int_0^T f(t)^2 dt\right)^{1/2}
\tag{24}\\
||f||_{L^1} &= \int_0^T |f(t)| dt
\tag{25}\\
||f||_{L^\infty} &= \max_{t\in [0,T]}|f(t)|
\tag{26}
\end{align}
$$
Norms of mesh functions
- Problem: \( f^n =f(t_n) \) is a mesh function and hence not defined for all \( t \). How to integrate \( f^n \)?
- Idea: Apply a numerical integration rule, using only the mesh points of the mesh function.
The Trapezoidal rule:
$$ ||f^n|| = \left(\Delta t\left(\half(f^0)^2 + \half(f^{N_t})^2
+ \sum_{n=1}^{N_t-1} (f^n)^2\right)\right)^{1/2} $$
Common simplification yields the \( L^2 \) norm of a mesh function:
$$ ||f^n||_{\ell^2} = \left(\Delta t\sum_{n=0}^{N_t} (f^n)^2\right)^{1/2} \tp$$
Implementation of the norm of the error
$$ E = ||e^n||_{\ell^2} = \sqrt{\Delta t\sum_{n=0}^{N_t} (e^n)^2}$$
Python w/array arithmetics:
e = u_exact(t) - u
E = sqrt(dt*sum(e**2))
Comment on array vs scalar computation
Scalar computing of E = sqrt(dt*sum(e**2)):
m = len(u) # length of u array (alt: u.size)
u_e = zeros(m)
t = 0
for i in range(m):
u_e[i] = exact_solution(t, a, I)
t = t + dt
e = zeros(m)
for i in range(m):
e[i] = u_e[i] - u[i]
s = 0 # summation variable
for i in range(m):
s = s + e[i]**2
error = sqrt(dt*s)
Obviously, scalar computing
- takes more code
- is less readable
- runs much slower
Plotting solutions
Basic plotting with Matplotlib is much like MATLAB plotting
from matplotlib.pyplot import *
plot(t, u)
show()
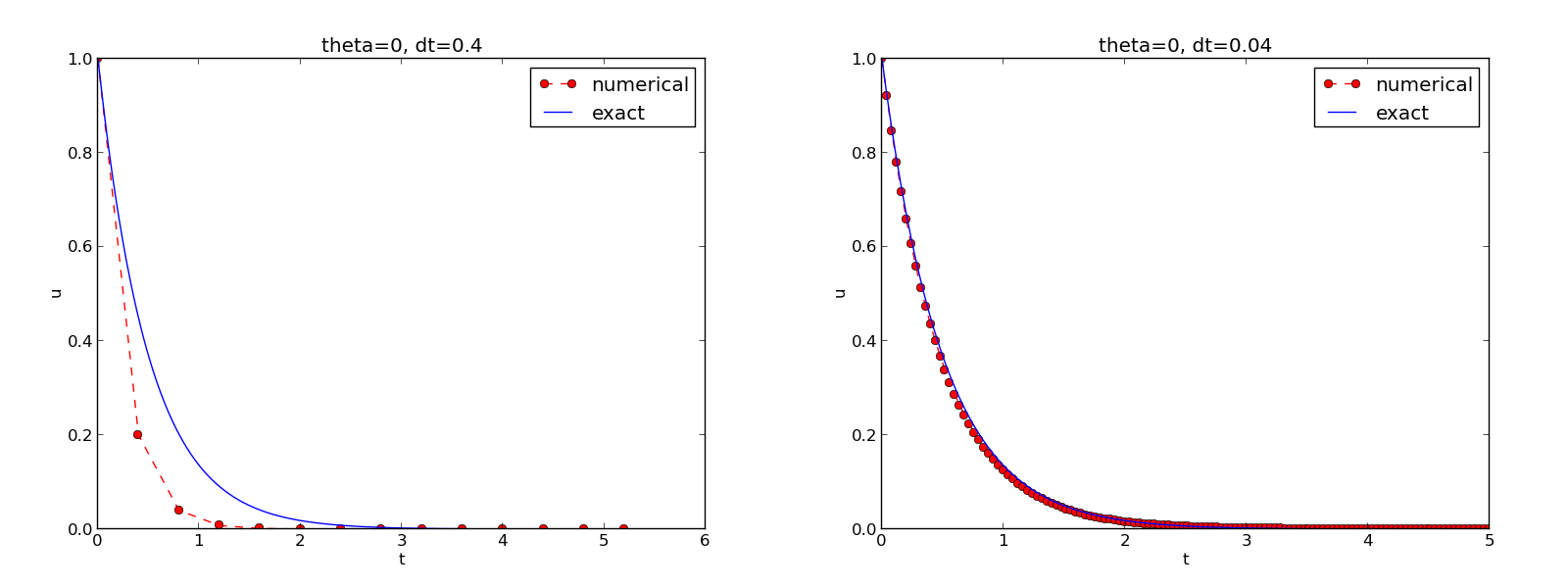
Compare u curve with \( \uex(t) \):
t_e = linspace(0, T, 1001) # fine mesh
u_e = exact_solution(t_e, I, a)
plot(t_e, u_e, 'b-') # blue line for u_e
plot(t, u, 'r--o') # red dashes w/circles

Decorating a plot
- Use different line types
- Add axis labels
- Add curve legends
- Add plot title
- Save plot to file
from matplotlib.pyplot import *
figure() # create new plot
t_e = linspace(0, T, 1001) # fine mesh for u_e
u_e = exact_solution(t_e, I, a)
plot(t, u, 'r--o') # red dashes w/circles
plot(t_e, u_e, 'b-') # blue line for exact sol.
legend(['numerical', 'exact'])
xlabel('t')
ylabel('u')
title('theta=%g, dt=%g' % (theta, dt))
savefig('%s_%g.png' % (theta, dt))
show()
See complete code in decay_plot_mpl.py.
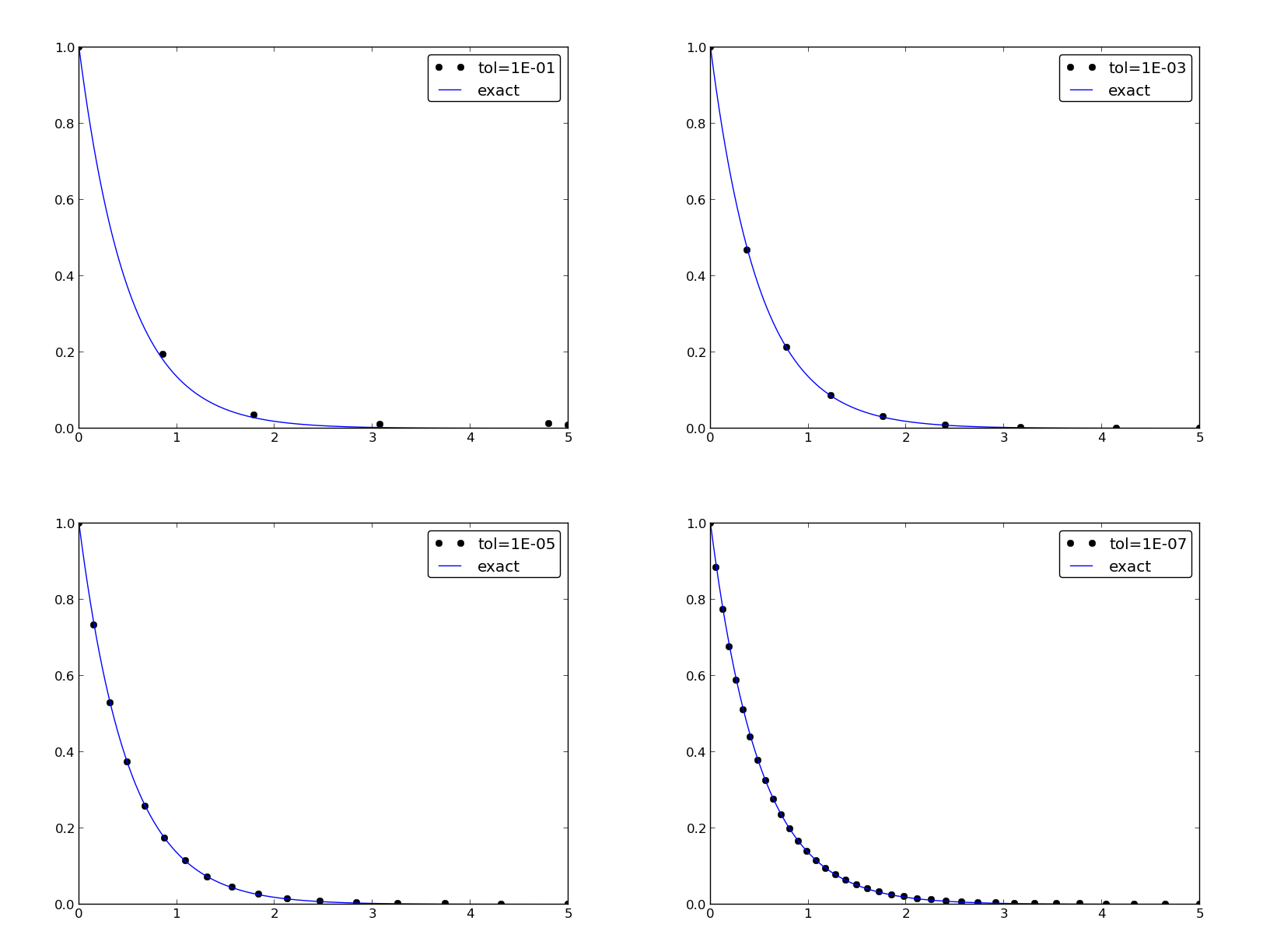
How the plots look like
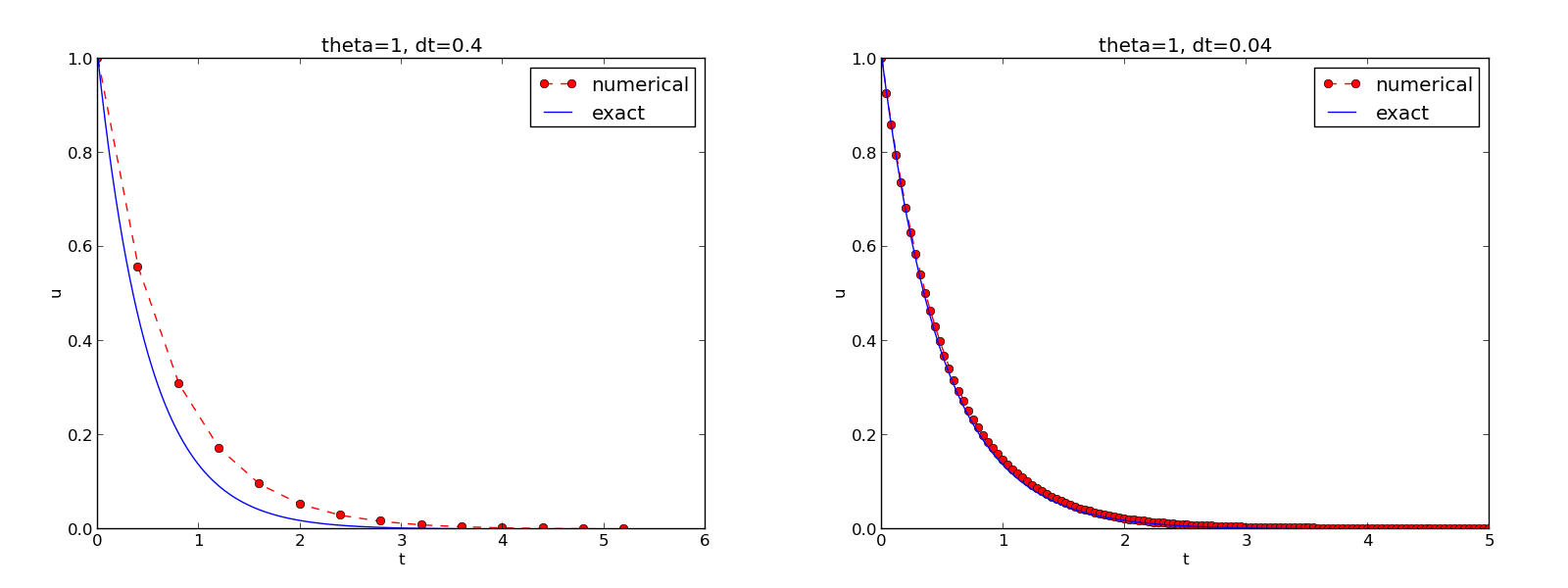
Plotting with SciTools
SciTools provides a unified plotting interface (Easyviz) to many different plotting packages: Matplotlib, Gnuplot, Grace, VTK, OpenDX, ...
Can use Matplotlib (MATLAB-like) syntax,
or a more compact plot function syntax:
from scitools.std import *
plot(t, u, 'r--o', # red dashes w/circles
t_e, u_e, 'b-', # blue line for exact sol.
legend=['numerical', 'exact'],
xlabel='t',
ylabel='u',
title='theta=%g, dt=%g' % (theta, dt),
savefig='%s_%g.png' % (theta2name[theta], dt),
show=True)
Complete code in decay_plot_st.py.
Change backend (plotting engine, Matplotlib by default):
Terminal> python decay_plot_st.py --SCITOOLS_easyviz_backend gnuplot
Terminal> python decay_plot_st.py --SCITOOLS_easyviz_backend grace
Creating user interfaces
- Never edit the program to change input!
- Set input data on the command line or in a graphical user interface
- How is explained next
Accessing command-line arguments
- All command-line arguments are available in
sys.argv -
sys.argv[0]is the program -
sys.argv[1:]holds the command-line arguments - Method 1: fixed sequence of parameters on the command line
- Method 2:
--option valuepairs on the command line (with default values)
Terminal> python myprog.py 1.5 2 0.5 0.8 0.4
Terminal> python myprog.py --I 1.5 --a 2 --dt 0.8 0.4
Reading a sequence of command-line arguments
The program decay_plot_mpl.py needs this input:
- \( I \)
- \( a \)
- \( T \)
- an option to turn the plot on or off (
makeplot) - a list of \( \Delta t \) values
Give these on the command line in correct sequence
Terminal> python decay_cml.py 1.5 2 0.5 0.8 0.4
Implementation
import sys
def read_command_line():
if len(sys.argv) < 6:
print 'Usage: %s I a T on/off dt1 dt2 dt3 ...' % \
sys.argv[0]; sys.exit(1) # abort
I = float(sys.argv[1])
a = float(sys.argv[2])
T = float(sys.argv[3])
makeplot = sys.argv[4] in ('on', 'True')
dt_values = [float(arg) for arg in sys.argv[5:]]
return I, a, T, makeplot, dt_values
Note:
-
sys.argv[i]is always a string - Must explicitly convert to (e.g.)
floatfor computations - List comprehensions make lists:
[expression for e in somelist]
Complete program: decay_cml.py.
Working with an argument parser
Set option-value pairs on the command line if the default value is not suitable:
Terminal> python decay_argparse.py --I 1.5 --a 2 --dt 0.8 0.4
Code:
def define_command_line_options():
import argparse
parser = argparse.ArgumentParser()
parser.add_argument('--I', '--initial_condition', type=float,
default=1.0, help='initial condition, u(0)',
metavar='I')
parser.add_argument('--a', type=float,
default=1.0, help='coefficient in ODE',
metavar='a')
parser.add_argument('--T', '--stop_time', type=float,
default=1.0, help='end time of simulation',
metavar='T')
parser.add_argument('--makeplot', action='store_true',
help='display plot or not')
parser.add_argument('--dt', '--time_step_values', type=float,
default=[1.0], help='time step values',
metavar='dt', nargs='+', dest='dt_values')
return parser
(metavar is the symbol used in help output)
Reading option-values pairs
argparse.ArgumentParser parses the command-line arguments:
def read_command_line():
parser = define_command_line_options()
args = parser.parse_args()
print 'I={}, a={}, T={}, makeplot={}, dt_values={}'.format(
args.I, args.a, args.T, args.makeplot, args.dt_values)
return args.I, args.a, args.T, args.makeplot, args.dt_values
Complete program: decay_argparse.py.
A graphical user interface
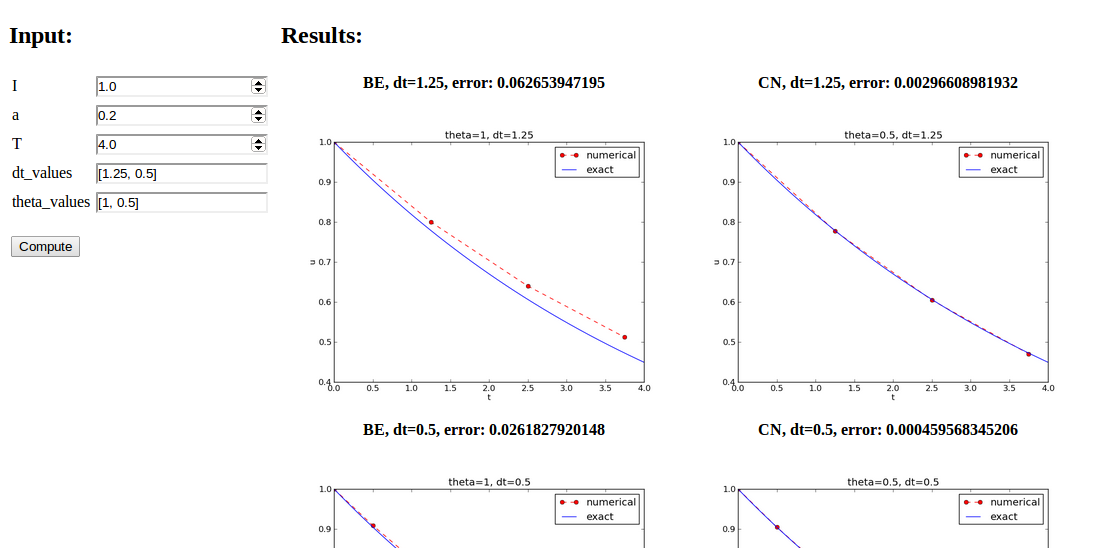
Normally very much programming required - and much competence on graphical user interfaces.
Here: use a tool to automatically create it in a few minutes (!)
The Parampool package
- Parampool is a package for handling a large pool of input parameters in simulation programs
- Parampool can automatically create a sophisticated web-based graphical user interface (GUI) to set parameters and view solutions
Making a compute function
- Key concept: a compute function that takes all input data as arguments and returning HTML code for viewing the results (e.g., plots and numbers)
- What we have: decay_plot_mpl.py
-
mainfunction carries out simulations and plotting for a series of \( \Delta t \) values - Goal: steer and view these experiments from a web GUI
- What to do:
- create a compute function
- call
parampoolfunctionality
The compute function main_GUI:
def main_GUI(I=1.0, a=.2, T=4.0,
dt_values=[1.25, 0.75, 0.5, 0.1],
theta_values=[0, 0.5, 1]):
The hard part of the compute function: the HTML code
- The results are to be displayed in a web page
- Only you know what to display in your problem
- Therefore, you need to specify the HTML code
Suppose explore solves the problem, makes a plot, computes the
error and returns appropriate HTML code with the plot. Embed
error and plots in a table:
def main_GUI(I=1.0, a=.2, T=4.0,
dt_values=[1.25, 0.75, 0.5, 0.1],
theta_values=[0, 0.5, 1]):
# Build HTML code for web page. Arrange plots in columns
# corresponding to the theta values, with dt down the rows
theta2name = {0: 'FE', 1: 'BE', 0.5: 'CN'}
html_text = '<table>\n'
for dt in dt_values:
html_text += '<tr>\n'
for theta in theta_values:
E, html = explore(I, a, T, dt, theta, makeplot=True)
html_text += """
<td>
<center><b>%s, dt=%g, error: %s</b></center><br>
%s
</td>
""" % (theta2name[theta], dt, E, html)
html_text += '</tr>\n'
html_text += '</table>\n'
return html_text
How to embed a PNG plot in HTML code
In explore:
import matplotlib.pyplot as plt
...
# plot
plt.plot(t, u, r-')
plt.xlabel('t')
plt.ylabel('u')
...
from parampool.utils import save_png_to_str
html_text = save_png_to_str(plt, plotwidth=400)
If you know HTML, you can return more sophisticated layout etc.
Generating the user interface
Make a file decay_GUI_generate.py:
from parampool.generator.flask import generate
from decay_GUI import main
generate(main,
output_controller='decay_GUI_controller.py',
output_template='decay_GUI_view.py',
output_model='decay_GUI_model.py')
Running decay_GUI_generate.py results in
-
decay_GUI_model.pydefines HTML widgets to be used to set input data in the web interface, -
templates/decay_GUI_views.pydefines the layout of the web page, -
decay_GUI_controller.pyruns the web application.
Good news: we only need to run decay_GUI_controller.py
and there is no need to look into any of these files!
Running the web application
Start the GUI
Terminal> python decay_GUI_controller.py
Open a web browser at 127.0.0.1:5000

More advanced use
- The compute function can have arguments of type float, int, string, list, dict, numpy array, filename (file upload)
- Alternative: specify a hierarchy of input parameters with name, default value, data type, widget type, unit (m, kg, s), validity check
- The generated web GUI can have user accounts with login and storage of results in a database
Computing convergence rates
Frequent assumption on the relation between the numerical error \( E \) and some discretization parameter \( \Delta t \):
$$
\begin{equation}
E = C\Delta t^r,
\tag{27}
\end{equation}
$$
- Unknown: \( C \) and \( r \).
- Goal: estimate \( r \) (and \( C \)) from numerical experiments
Estimating the convergence rate \( r \)
Perform numerical experiments: \( (\Delta t_i, E_i) \), \( i=0,\ldots,m-1 \). Two methods for finding \( r \) (and \( C \)):
- Take the logarithm of (27), \( \ln E = r\ln \Delta t + \ln C \), and fit a straight line to the data points \( (\Delta t_i, E_i) \), \( i=0,\ldots,m-1 \).
- Consider two consecutive experiments, \( (\Delta t_i, E_i) \) and \( (\Delta t_{i-1}, E_{i-1}) \). Dividing the equation \( E_{i-1}=C\Delta t_{i-1}^r \) by \( E_{i}=C\Delta t_{i}^r \) and solving for \( r \) yields
$$
\begin{equation}
r_{i-1} = \frac{\ln (E_{i-1}/E_i)}{\ln (\Delta t_{i-1}/\Delta t_i)}
\tag{28}
\end{equation}
$$
for \( i=1,=\ldots,m-1 \).
Method 2 is best.
Implementation
Compute \( r_0, r_1, \ldots, r_{m-2} \):
from math import log
def main():
I, a, T, makeplot, dt_values = read_command_line()
r = {} # estimated convergence rates
for theta in 0, 0.5, 1:
E_values = []
for dt in dt_values:
E = explore(I, a, T, dt, theta, makeplot=False)
E_values.append(E)
# Compute convergence rates
m = len(dt_values)
r[theta] = [log(E_values[i-1]/E_values[i])/
log(dt_values[i-1]/dt_values[i])
for i in range(1, m, 1)]
for theta in r:
print '\nPairwise convergence rates for theta=%g:' % theta
print ' '.join(['%.2f' % r_ for r_ in r[theta]])
return r
Complete program: decay_convrate.py.
Execution
Terminal> python decay_convrate.py --dt 0.5 0.25 0.1 0.05 0.025 0.01
...
Pairwise convergence rates for theta=0:
1.33 1.15 1.07 1.03 1.02
Pairwise convergence rates for theta=0.5:
2.14 2.07 2.03 2.01 2.01
Pairwise convergence rates for theta=1:
0.98 0.99 0.99 1.00 1.00
Debugging via convergence rates
Potential bug: missing a in the denominator,
u[n+1] = (1 - (1-theta)*a*dt)/(1 + theta*dt)*u[n]
Running decay_convrate.py gives same rates.
Why? The value of \( a \)... (\( a=1 \))
0 and 1 are bad values in tests!
Better:
Terminal> python decay_convrate.py --a 2.1 --I 0.1 \
--dt 0.5 0.25 0.1 0.05 0.025 0.01
...
Pairwise convergence rates for theta=0:
1.49 1.18 1.07 1.04 1.02
Pairwise convergence rates for theta=0.5:
-1.42 -0.22 -0.07 -0.03 -0.01
Pairwise convergence rates for theta=1:
0.21 0.12 0.06 0.03 0.01
Forward Euler works...because \( \theta=0 \) hides the bug.
This bug gives \( r\approx 0 \):
u[n+1] = ((1-theta)*a*dt)/(1 + theta*dt*a)*u[n]
Memory-saving implementation
- Note 1: we store the entire array
u, i.e., \( u^n \) for \( n=0,1,\ldots,N_t \) - Note 2: the formula for \( u^{n+1} \) needs \( u^n \) only, not \( u^{n-1} \), \( u^{n-2} \), ...
- No need to store more than \( u^{n+1} \) and \( u^{n} \)
- Extremely important when solving PDEs
- No practical importance here (much memory available)
- But let's illustrate how to do save memory!
- Idea 1: store \( u^{n+1} \) in
u, \( u^n \) inu_1(float) - Idea 2: store
uin a file, read file later for plotting
Memory-saving solver function
def solver_memsave(I, a, T, dt, theta, filename='sol.dat'):
"""
Solve u'=-a*u, u(0)=I, for t in (0,T] with steps of dt.
Minimum use of memory. The solution is stored in a file
(with name filename) for later plotting.
"""
dt = float(dt) # avoid integer division
Nt = int(round(T/dt)) # no of intervals
outfile = open(filename, 'w')
# u: time level n+1, u_1: time level n
t = 0
u_1 = I
outfile.write('%.16E %.16E\n' % (t, u_1))
for n in range(1, Nt+1):
u = (1 - (1-theta)*a*dt)/(1 + theta*dt*a)*u_1
u_1 = u
t += dt
outfile.write('%.16E %.16E\n' % (t, u))
outfile.close()
return u, t
Reading computed data from file
def read_file(filename='sol.dat'):
infile = open(filename, 'r')
u = []; t = []
for line in infile:
words = line.split()
if len(words) != 2:
print 'Found more than two numbers on a line!', words
sys.exit(1) # abort
t.append(float(words[0]))
u.append(float(words[1]))
return np.array(t), np.array(u)
Simpler code with numpy functionality for reading/writing tabular data:
def read_file_numpy(filename='sol.dat'):
data = np.loadtxt(filename)
t = data[:,0]
u = data[:,1]
return t, u
Similar function np.savetxt, but then we need all \( u^n \) and \( t^n \) values
in a two-dimensional array (which we try to prevent now!).
Usage of memory-saving code
def explore(I, a, T, dt, theta=0.5, makeplot=True):
filename = 'u.dat'
u, t = solver_memsave(I, a, T, dt, theta, filename)
t, u = read_file(filename)
u_e = exact_solution(t, I, a)
e = u_e - u
E = np.sqrt(dt*np.sum(e**2))
if makeplot:
plt.figure()
...
Complete program: decay_memsave.py.
Software engineering
Goal: make more professional numerical software.
Topics:
- How to make modules (reusable libraries)
- Testing frameworks (doctest, nose, unittest)
- Implementation with classes
Making a module
- Previous programs: much repetitive code (esp.
solver) - DRY (Don't Repeat Yourself) principle: no copies of code
- A change needs to be done in one and only one place
- Module = just a file with functions (reused through
import) - Let's make a module by putting these functions in a file:
-
solver -
verify_three_steps -
verify_discrete_solution -
explore -
define_command_line_options -
read_command_line -
main(with convergence rates) -
verify_convergence_rate
Module name: decay_mod, filename: decay_mod.py.
Sketch:
from numpy import *
from matplotlib.pyplot import *
import sys
def solver(I, a, T, dt, theta):
...
def verify_three_steps():
...
def verify_exact_discrete_solution():
...
def exact_solution(t, I, a):
...
def explore(I, a, T, dt, theta=0.5, makeplot=True):
...
def define_command_line_options():
...
def read_command_line(use_argparse=True):
...
def main():
...
That is! It's a module decay_mod in file decay_mod.py.
Usage in some other program:
from decay_mod import solver
u, t = solver(I=1.0, a=3.0, T=3, dt=0.01, theta=0.5)
Test block
At the end of a module it is common to include a test block:
if __name__ == '__main__':
main()
- If
decay_modis imported,__name__isdecay_mod. - If
decay_mod.pyis run,__name__is__main__. - Use test block for testing, demo, user interface, ...
Extended test block:
if __name__ == '__main__':
if 'verify' in sys.argv:
if verify_three_steps() and verify_discrete_solution():
pass # ok
else:
print 'Bug in the implementation!'
elif 'verify_rates' in sys.argv:
sys.argv.remove('verify_rates')
if not '--dt' in sys.argv:
print 'Must assign several dt values'
sys.exit(1) # abort
if verify_convergence_rate():
pass
else:
print 'Bug in the implementation!'
else:
# Perform simulations
main()
Prefixing imported functions by the module name
from numpy import *
from matplotlib.pyplot import *
This imports a large number of names (sin, exp, linspace, plot, ...).
Confusion: is a function from`numpy`? Or matplotlib.pyplot?
Alternative (recommended) import:
import numpy
import matplotlib.pyplot
Now we need to prefix functions with module name:
t = numpy.linspace(0, T, Nt+1)
u_e = I*numpy.exp(-a*t)
matplotlib.pyplot.plot(t, u_e)
Common standard:
import numpy as np
import matplotlib.pyplot as plt
t = np.linspace(0, T, Nt+1)
u_e = I*np.exp(-a*t)
plt.plot(t, u_e)
Downside of module prefix notation
A math line like \( e^{-at}\sin(2\pi t) \) gets cluttered with module names,
numpy.exp(-a*t)*numpy.sin(2(numpy.pi*t)
# or
np.exp(-a*t)*np.sin(2*np.pi*t)
Solution (much used in this course): do two imports
import numpy as np
from numpy import exp, sin, pi
...
t = np.linspace(0, T, Nt+1)
u_e = exp(-a*t)*sin(2*pi*t)
Doctests
Doc strings can be equipped with interactive Python sessions for demonstrating usage and automatic testing of functions.
def solver(I, a, T, dt, theta):
"""
Solve u'=-a*u, u(0)=I, for t in (0,T] with steps of dt.
>>> u, t = solver(I=0.8, a=1.2, T=4, dt=0.5, theta=0.5)
>>> for t_n, u_n in zip(t, u):
... print 't=%.1f, u=%.14f' % (t_n, u_n)
t=0.0, u=0.80000000000000
t=0.5, u=0.43076923076923
t=1.0, u=0.23195266272189
t=1.5, u=0.12489758761948
t=2.0, u=0.06725254717972
t=2.5, u=0.03621291001985
t=3.0, u=0.01949925924146
t=3.5, u=0.01049960113002
t=4.0, u=0.00565363137770
"""
...
Running doctests
Automatic check that the code reproduces the doctest output:
Terminal> python -m doctest decay_mod_doctest.py
Report in case of failure:
Terminal> python -m doctest decay_mod_doctest.py
********************************************************
File "decay_mod_doctest.py", line 12, in decay_mod_doctest....
Failed example:
for t_n, u_n in zip(t, u):
print 't=%.1f, u=%.14f' % (t_n, u_n)
Expected:
t=0.0, u=0.80000000000000
t=0.5, u=0.43076923076923
t=1.0, u=0.23195266272189
t=1.5, u=0.12489758761948
t=2.0, u=0.06725254717972
Got:
t=0.0, u=0.80000000000000
t=0.5, u=0.43076923076923
t=1.0, u=0.23195266272189
t=1.5, u=0.12489758761948
t=2.0, u=0.06725254718756
********************************************************
1 items had failures:
1 of 2 in decay_mod_doctest.solver
***Test Failed*** 1 failures.
Complete program: decay_mod_doctest.py.
Unit testing with nose
- Nose is a very user-friendly testing framework
- Based on unit testing
- Identify (small) units of code and test each unit
- Nose automates running all tests
- Good habit: run all tests after (small) edits of a code
- Even better habit: write tests before the code (!)
- Remark: unit testing in scientific computing is not yet well established
Basic use of nose
- Implement tests in test functions with names starting with
test_. - Test functions cannot have arguments.
- Test functions perform assertions on computed results
using
assertfunctions from thenose.toolsmodule. - Test functions can be in the source code files or be
collected in separate files
test*.py.
Example on a nose test in the source code
Very simple module mymod (in file mymod.py):
def double(n):
return 2*n
Write test function in mymod.py:
def double(n):
return 2*n
import nose.tools as nt
def test_double():
result = double(4)
nt.assert_equal(result, 8)
Running
Terminal> nosetests -s mymod
makes the nose tool run all test_*() functions in mymod.py.
Example on a nose test in a separate file
Write the test in a separate file, say test_mymod.py:
import nose.tools as nt
import mymod
def test_double():
result = mymod.double(4)
nt.assert_equal(result, 8)
Running
Terminal> nosetests -s
makes the nose tool run all test_*() functions in all files
test*.py in the current directory and in all subdirectories (recursevely)
with names tests or *_tests.
The habit of writing nose tests
- Put
test_*()functions in the module - When you get many
test_*()functions, collect them intests/test*.py
Purpose of a test function: raise AssertionError if failure
Alternative ways of raising AssertionError if result is not 8:
import nose.tools as nt
def test_double():
result = ...
nt.assert_equal(result, 8) # alternative 1
assert result == 8 # alternative 2
if result != 8: # alternative 3
raise AssertionError()
Advantages of nose
- Easier to use than other test frameworks
- Tests are written and collected in a compact and structured way
- Large collections of tests, scattered throughout a directory tree
can be executed with one command (
nosetests -s) - Nose is a much-adopted standard
Demonstrating nose (ideas)
Aim: test function solver for \( u'=-au \), \( u(0)=I \).
We design three unit tests:
- A comparison between the computed \( u^n \) values and the exact discrete solution
- A comparison between the computed \( u^n \) values and precomputed verified reference values
- A comparison between observed and expected convergence rates
These tests follow very closely the previous verify* functions.
Demonstrating nose (code)
import nose.tools as nt
import decay_mod_unittest as decay_mod
import numpy as np
def exact_discrete_solution(n, I, a, theta, dt):
"""Return exact discrete solution of the theta scheme."""
dt = float(dt) # avoid integer division
factor = (1 - (1-theta)*a*dt)/(1 + theta*dt*a)
return I*factor**n
def test_exact_discrete_solution():
"""
Compare result from solver against
formula for the discrete solution.
"""
theta = 0.8; a = 2; I = 0.1; dt = 0.8
N = int(8/dt) # no of steps
u, t = decay_mod.solver(I=I, a=a, T=N*dt, dt=dt, theta=theta)
u_de = np.array([exact_discrete_solution(n, I, a, theta, dt)
for n in range(N+1)])
diff = np.abs(u_de - u).max()
nt.assert_almost_equal(diff, 0, delta=1E-14)
Floats as test results require careful comparison
- Round-off errors make exact comparison of floats unreliable
-
nt.assert_almost_equal: compare two floats to some digits or precision
def test_solver():
"""
Compare result from solver against
precomputed arrays for theta=0, 0.5, 1.
"""
I=0.8; a=1.2; T=4; dt=0.5 # fixed parameters
precomputed = {
't': np.array([ 0. , 0.5, 1. , 1.5, 2. , 2.5,
3. , 3.5, 4. ]),
0.5: np.array(
[ 0.8 , 0.43076923, 0.23195266, 0.12489759,
0.06725255, 0.03621291, 0.01949926, 0.0104996 ,
0.00565363]),
0: ...,
1: ...
}
for theta in 0, 0.5, 1:
u, t = decay_mod.solver(I, a, T, dt, theta=theta)
diff = np.abs(u - precomputed[theta]).max()
# Precomputed numbers are known to 8 decimal places
nt.assert_almost_equal(diff, 0, places=8,
msg='theta=%s' % theta)
Test of wrong use
- Find input data that may cause trouble and test such cases
- Here: the formula for \( u^{n+1} \) may involve integer division
Example:
theta = 1; a = 1; I = 1; dt = 2
may lead to integer division:
(1 - (1-theta)*a*dt) # becomes 1
(1 + theta*dt*a) # becomes 2
(1 - (1-theta)*a*dt)/(1 + theta*dt*a) # becomes 0 (!)
Test that solver does not suffer from such integer division:
def test_potential_integer_division():
"""Choose variables that can trigger integer division."""
theta = 1; a = 1; I = 1; dt = 2
N = 4
u, t = decay_mod.solver(I=I, a=a, T=N*dt, dt=dt, theta=theta)
u_de = np.array([exact_discrete_solution(n, I, a, theta, dt)
for n in range(N+1)])
diff = np.abs(u_de - u).max()
nt.assert_almost_equal(diff, 0, delta=1E-14)
Test of convergence rates
Convergence rate tests are very common for differential equation solvers.
def test_convergence_rates():
"""Compare empirical convergence rates to exact ones."""
# Set command-line arguments directly in sys.argv
import sys
sys.argv[1:] = '--I 0.8 --a 2.1 --T 5 '\
'--dt 0.4 0.2 0.1 0.05 0.025'.split()
r = decay_mod.main()
for theta in r:
nt.assert_true(r[theta]) # check for non-empty list
expected_rates = {0: 1, 1: 1, 0.5: 2}
for theta in r:
r_final = r[theta][-1]
# Compare to 1 decimal place
nt.assert_almost_equal(expected_rates[theta], r_final,
places=1, msg='theta=%s' % theta)
Complete program: test_decay_nose.py.
Classical unit testing with unittest
-
unittestis a Python module mimicing the classical JUnit class-based unit testing framework from Java - This is how unit testing is normally done
- Requires knowledge of object-oriented programming
Basic use of unittest
Write file test_mymod.py:
import unittest
import mymod
class TestMyCode(unittest.TestCase):
def test_double(self):
result = mymod.double(4)
self.assertEqual(result, 8)
if __name__ == '__main__':
unittest.main()
Demonstration of unittest
import unittest
import decay_mod_unittest as decay
import numpy as np
def exact_discrete_solution(n, I, a, theta, dt):
factor = (1 - (1-theta)*a*dt)/(1 + theta*dt*a)
return I*factor**n
class TestDecay(unittest.TestCase):
def test_exact_discrete_solution(self):
...
diff = np.abs(u_de - u).max()
self.assertAlmostEqual(diff, 0, delta=1E-14)
def test_solver(self):
...
for theta in 0, 0.5, 1:
...
self.assertAlmostEqual(diff, 0, places=8,
msg='theta=%s' % theta)
def test_potential_integer_division():
...
self.assertAlmostEqual(diff, 0, delta=1E-14)
def test_convergence_rates(self):
...
for theta in r:
...
self.assertAlmostEqual(...)
if __name__ == '__main__':
unittest.main()
Complete program: test_decay_unittest.py.
Implementing simple problem and solver classes
- So far: programs are built of Python functions
- New focus: alternative implementations using classes
- Class-based implementations are very popular, especially in business/adm applications
- Class-based implementations scales better to large and complex scientific applications
What to learn
Tasks:
- Explain basic use of classes to build a differential equation solver
- Introduce concepts that make such programs easily scale to more complex applications
- Demonstrate the advantage of using classes
Ideas:
- Classes for Problem, Solver, and Visualizer
- Problem: all the physics information about the problem
- Solver: all the numerics information + numerical computations
- Visualizer: plot the solution and other quantities
The problem class
- Model problem: \( u'=-au \), \( u(0)=I \), for \( t\in (0,T] \).
- Class
Problemstores the physical parameters \( a \), \( I \), \( T \) - May also offer other data, e.g., \( \uex(t)=Ie^{-at} \)
Implementation:
from numpy import exp
class Problem:
def __init__(self, I=1, a=1, T=10):
self.T, self.I, self.a = I, float(a), T
def exact_solution(self, t):
I, a = self.I, self.a # extract local variables
return I*exp(-a*t)
Basic usage:
problem = Problem(T=5)
problem.T = 8
problem.dt = 1.5
Improved problem class
More flexible input from the command line:
class Problem:
def __init__(self, I=1, a=1, T=10):
self.T, self.I, self.a = I, float(a), T
def define_command_line_options(self, parser=None):
if parser is None:
import argparse
parser = argparse.ArgumentParser()
parser.add_argument(
'--I', '--initial_condition', type=float,
default=self.I, help='initial condition, u(0)',
metavar='I')
parser.add_argument(
'--a', type=float, default=self.a,
help='coefficient in ODE', metavar='a')
parser.add_argument(
'--T', '--stop_time', type=float, default=self.T,
help='end time of simulation', metavar='T')
return parser
def init_from_command_line(self, args):
self.I, self.a, self.T = args.I, args.a, args.T
def exact_solution(self, t):
I, a = self.I, self.a
return I*exp(-a*t)
- Can utilize user's
ArgumentParser, or make one -
Noneis used to indicate a non-initialized variable
The solver class
- Store numerical data \( \Delta t \), \( \theta \)
- Compute solution and quantities derived from the solution
Implementation:
class Solver:
def __init__(self, problem, dt=0.1, theta=0.5):
self.problem = problem
self.dt, self.theta = float(dt), theta
def define_command_line_options(self, parser):
parser.add_argument(
'--dt', '--time_step_value', type=float,
default=0.5, help='time step value', metavar='dt')
parser.add_argument(
'--theta', type=float, default=0.5,
help='time discretization parameter', metavar='dt')
return parser
def init_from_command_line(self, args):
self.dt, self.theta = args.dt, args.theta
def solve(self):
from decay_mod import solver
self.u, self.t = solver(
self.problem.I, self.problem.a, self.problem.T,
self.dt, self.theta)
Note: reuse of the numerical algorithm from the decay_mod module
(i.e., the class is a wrapper of the procedural implementation).
The visualizer class
class Visualizer:
def __init__(self, problem, solver):
self.problem, self.solver = problem, solver
def plot(self, include_exact=True, plt=None):
"""
Add solver.u curve to the plotting object plt,
and include the exact solution if include_exact is True.
This plot function can be called several times (if
the solver object has computed new solutions).
"""
if plt is None:
import scitools.std as plt # can use matplotlib as well
plt.plot(self.solver.t, self.solver.u, '--o')
plt.hold('on')
theta2name = {0: 'FE', 1: 'BE', 0.5: 'CN'}
name = theta2name.get(self.solver.theta, '')
legends = ['numerical %s' % name]
if include_exact:
t_e = linspace(0, self.problem.T, 1001)
u_e = self.problem.exact_solution(t_e)
plt.plot(t_e, u_e, 'b-')
legends.append('exact')
plt.legend(legends)
plt.xlabel('t')
plt.ylabel('u')
plt.title('theta=%g, dt=%g' %
(self.solver.theta, self.solver.dt))
plt.savefig('%s_%g.png' % (name, self.solver.dt))
return plt
Remark: The plt object in plot adds a new curve to a plot,
which enables comparing different solutions from different
runs of Solver.solve
Combing the classes
Let Problem, Solver, and Visualizer play together:
def main():
problem = Problem()
solver = Solver(problem)
viz = Visualizer(problem, solver)
# Read input from the command line
parser = problem.define_command_line_options()
parser = solver. define_command_line_options(parser)
args = parser.parse_args()
problem.init_from_command_line(args)
solver. init_from_command_line(args)
# Solve and plot
solver.solve()
import matplotlib.pyplot as plt
#import scitools.std as plt
plt = viz.plot(plt=plt)
E = solver.error()
if E is not None:
print 'Error: %.4E' % E
plt.show()
Complete program: decay_class.py.
Implementing more advanced problem and solver classes
- The previous
ProblemandSolverclasses soon contain much repetitive code when the number of parameters increases - Much of such code can be parameterized and be made more compact
- Idea: collect all parameters in a dictionary
self.prms, with two associated dictionariesself.typesandself.helpfor holding associated object types and help strings - Collect common code in class
Parameters - Let
Problem,Solver, and maybeVisualizerbe subclasses of classParameters, basically definingself.prms,self.types,self.help
A generic class for parameters
class Parameters:
def set(self, **parameters):
for name in parameters:
self.prms[name] = parameters[name]
def get(self, name):
return self.prms[name]
def define_command_line_options(self, parser=None):
if parser is None:
import argparse
parser = argparse.ArgumentParser()
for name in self.prms:
tp = self.types[name] if name in self.types else str
help = self.help[name] if name in self.help else None
parser.add_argument(
'--' + name, default=self.get(name), metavar=name,
type=tp, help=help)
return parser
def init_from_command_line(self, args):
for name in self.prms:
self.prms[name] = getattr(args, name)
Slightly more advanced version in class_decay_verf1.py.
The problem class
class Problem(Parameters):
"""
Physical parameters for the problem u'=-a*u, u(0)=I,
with t in [0,T].
"""
def __init__(self):
self.prms = dict(I=1, a=1, T=10)
self.types = dict(I=float, a=float, T=float)
self.help = dict(I='initial condition, u(0)',
a='coefficient in ODE',
T='end time of simulation')
def exact_solution(self, t):
I, a = self.get('I'), self.get('a')
return I*np.exp(-a*t)
The solver class
class Solver(Parameters):
def __init__(self, problem):
self.problem = problem
self.prms = dict(dt=0.5, theta=0.5)
self.types = dict(dt=float, theta=float)
self.help = dict(dt='time step value',
theta='time discretization parameter')
def solve(self):
from decay_mod import solver
self.u, self.t = solver(
self.problem.get('I'),
self.problem.get('a'),
self.problem.get('T'),
self.get('dt'),
self.get('theta'))
def error(self):
try:
u_e = self.problem.exact_solution(self.t)
e = u_e - self.u
E = np.sqrt(self.get('dt')*np.sum(e**2))
except AttributeError:
E = None
return E
The visualizer class
- No parameters needed (for this simple problem), no need to inherit
class
Parameters - Same code as previously shown class
Visualizer - Same code as previously shown for combining
Problem,Solver, andVisualizer
Performing scientific experiments
Goal: explore the behavior of a numerical method for a differential equation and show how scientific experiments can be set up and reported.
Tasks:
- Write scripts to automate experiments
- Generate scientific reports from scripts
Tools to learn:
-
os.systemfor running other programs -
subprocessfor running other programs and extracting the output - List comprehensions
- Formats for scientific reports: HTML w/MathJax, LaTeX, Sphinx, Doconce
Model problem and numerical solution method
Problem:
$$
\begin{equation}
u'(t) = -au(t),\quad u(0)=I,\ 0< t \leq T,
\tag{29}
\end{equation}
$$
Solution method (\( \theta \)-rule):
$$
u^{n+1} = \frac{1 - (1-\theta) a\Delta t}{1 + \theta a\Delta t}u^n,
\quad u^0=I\tp
$$
Plan for the experiments
- Plot \( u^n \) against \( \uex = Ie^{-at} \) for various choices of the parameters \( I \), \( a \), \( \Delta t \), and \( \theta \)
- How does the discrete solution compare with the exact solution when \( \Delta t \) is varied and \( \theta=0,0.5,1 \)?
- Use the decay_mod.py module (little modification of the plotting, see experiments/decay_mod.py)
- Make separate program for running (automating) the experiments (script)
-
python decay_mod.py --I 1 --a 2 --makeplot --T 5 --dt 0.5 0.25 0.1 0.05 - Combine generated figures
FE_*.png,BE_*.png, andCN_*.pngto new figures with multiple plots - Run script as
python decay_exper0.py 0.5 0.25 0.1 0.05(\( \Delta t \) values on the command line)
Typical plot summarizing the results

Script code
Typical script (small administering program) for running the experiments:
import os, sys
def run_experiments(I=1, a=2, T=5):
# The command line must contain dt values
if len(sys.argv) > 1:
dt_values = [float(arg) for arg in sys.argv[1:]]
else:
print 'Usage: %s dt1 dt2 dt3 ...' % sys.argv[0]
sys.exit(1) # abort
# Run module file as a stand-alone application
cmd = 'python decay_mod.py --I %g --a %g --makeplot --T %g' % \
(I, a, T)
dt_values_str = ' '.join([str(v) for v in dt_values])
cmd += ' --dt %s' % dt_values_str
print cmd
failure = os.system(cmd)
if failure:
print 'Command failed:', cmd; sys.exit(1)
# Combine images into rows with 2 plots in each row
image_commands = []
for method in 'BE', 'CN', 'FE':
pdf_files = ' '.join(['%s_%g.pdf' % (method, dt)
for dt in dt_values])
png_files = ' '.join(['%s_%g.png' % (method, dt)
for dt in dt_values])
image_commands.append(
'montage -background white -geometry 100%' +
' -tile 2x %s %s.png' % (png_files, method))
image_commands.append(
'convert -trim %s.png %s.png' % (method, method))
image_commands.append(
'convert %s.png -transparent white %s.png' %
(method, method))
image_commands.append(
'pdftk %s output tmp.pdf' % pdf_files)
num_rows = int(round(len(dt_values)/2.0))
image_commands.append(
'pdfnup --nup 2x%d tmp.pdf' % num_rows)
image_commands.append(
'pdfcrop tmp-nup.pdf %s.pdf' % method)
for cmd in image_commands:
print cmd
failure = os.system(cmd)
if failure:
print 'Command failed:', cmd; sys.exit(1)
# Remove the files generated above and by decay_mod.py
from glob import glob
filenames = glob('*_*.png') + glob('*_*.pdf') + \
glob('*_*.eps') + glob('tmp*.pdf')
for filename in filenames:
os.remove(filename)
if __name__ == '__main__':
run_experiments()
Complete program: experiments/decay_exper0.py.
Comments to the code
Many useful constructs in the previous script:
-
[float(arg) for arg in sys.argv[1:]]builds a list of real numbers from all the command-line arguments -
failure = os.system(cmd)runs an operating system command (e.g., another program) -
sys.exit(1)aborts the program -
['%s_%s.png' % (method, dt) for dt in dt_values]builds a list of filenames from a list of numbers (dt_values) - All
montagecommands for creating composite figures are stored in a list and thereafter executed in a loop -
glob.glob('*_*.png')returns a list of the names of all files in the current folder where the filename matches the Unix wildcard notation*_*.png(meaning "any text, underscore, any text, and then `.png`") -
os.remove(filename)removes the file with namefilename
Interpreting output from other programs
In decay_exper0.py we run a program (os.system) and
want to grab the output, e.g.,
Terminal> python decay_plot_mpl.py
0.0 0.40: 2.105E-01
0.0 0.04: 1.449E-02
0.5 0.40: 3.362E-02
0.5 0.04: 1.887E-04
1.0 0.40: 1.030E-01
1.0 0.04: 1.382E-02
Tasks:
- read the output from the
decay_mod.pyprogram - interpret this output and store the \( E \) values in arrays for each \( \theta \) value
- plot \( E \) versus \( \Delta t \), for each \( \theta \), in a log-log plot
Code for grabbing output from another program
Use the subprocess module to grab output:
from subprocess import Popen, PIPE, STDOUT
p = Popen(cmd, shell=True, stdout=PIPE, stderr=STDOUT)
output, dummy = p.communicate()
failure = p.returncode
if failure:
print 'Command failed:', cmd; sys.exit(1)
Code for interpreting the grabbed output
- Run through the
outputstring, line by line - If the current line prints \( \theta \), \( \Delta t \), and \( E \), split the line into these three pieces and store the data
- Store data in a dictionary
errorswith keysdtand the three \( \theta \) values
errors = {'dt': dt_values, 1: [], 0: [], 0.5: []}
for line in output.splitlines():
words = line.split()
if words[0] in ('0.0', '0.5', '1.0'): # line with E?
# typical line: 0.0 1.25: 7.463E+00
theta = float(words[0])
E = float(words[2])
errors[theta].append(E)
Next: plot \( E \) versus \( \Delta t \) for \( \theta=0,0.5,1 \)
Complete program: experiments/decay_exper1.py. Fine recipe for
- how to run other programs
- how to extract and interpret output from other programs
- how to automate many manual steps in creating simulations and figures
Making a report
- Scientific investigations are best documented in a report!
- A sample report
- How can we write such a report?
- First problem: what format should I write in?
- Plain HTML, generated by decay_exper1_html.py
- HTML with MathJax, generated by decay_exper1_mathjax.py
- LaTeX PDF, based on LaTeX source
- Sphinx HTML, based on reStructuredText
- Markdown, MediaWiki, ...
- Doconce can generate LaTeX, HTML w/MathJax, Sphinx, Markdown, MediaWiki, ... (Doconce source for the examples above, and Python program for generating the Doconce source)
- Examples on different report formats
Publishing a complete project
- Make folder (directory) tree
- Keep track of all files via a version control system (Mercurial, Git, ...)
- Publish as private or public repository
- Utilize Bitbucket, Googlecode, GitHub, or similar
- See the intro to such tools
Analysis of finite difference equations
Model:
$$
\begin{equation}
u'(t) = -au(t),\quad u(0)=I,
\end{equation}
$$
Method:
$$
\begin{equation}
u^{n+1} = \frac{1 - (1-\theta) a\Delta t}{1 + \theta a\Delta t}u^n
\tag{30}
\end{equation}
$$
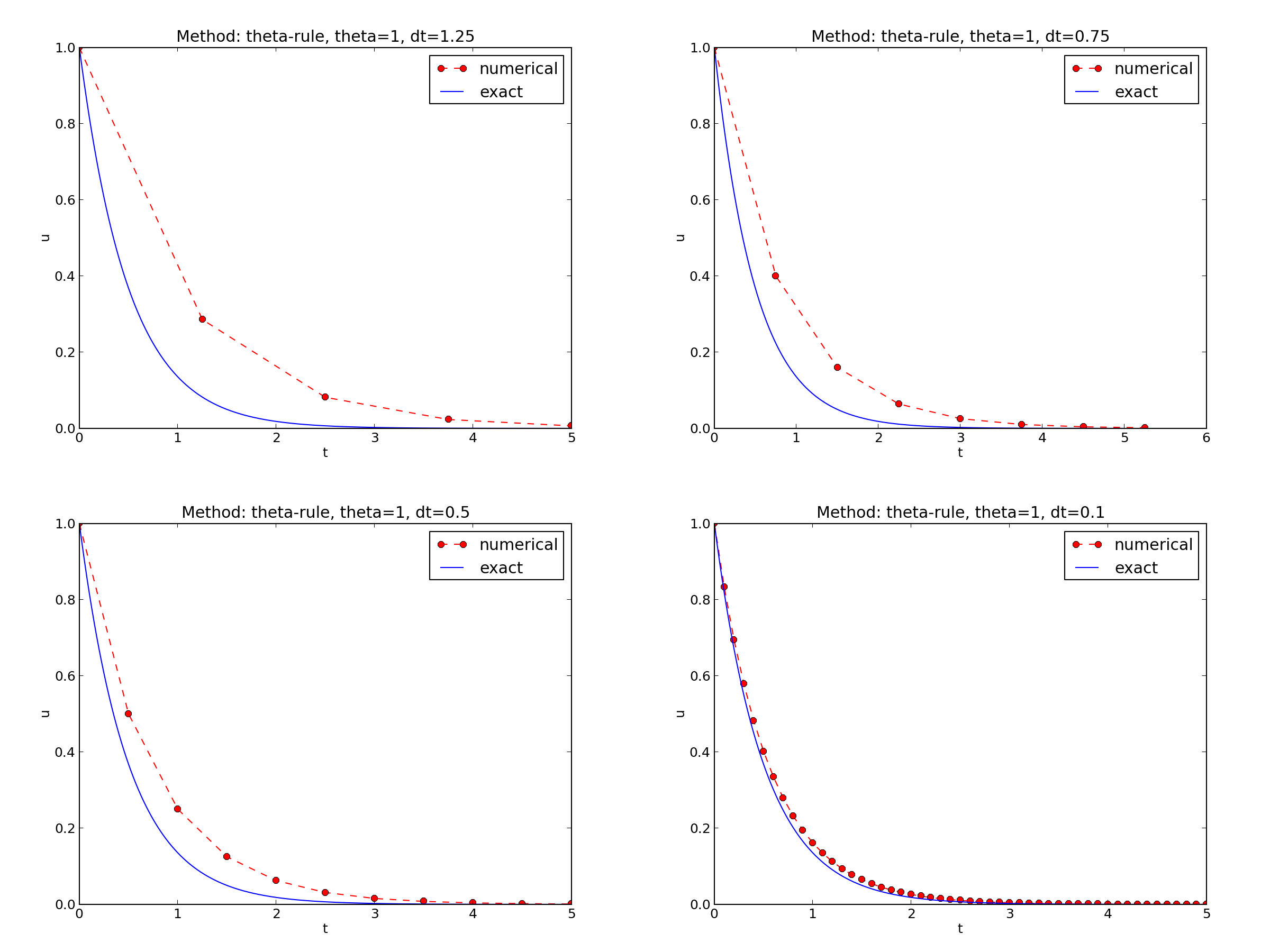
Encouraging numerical solutions
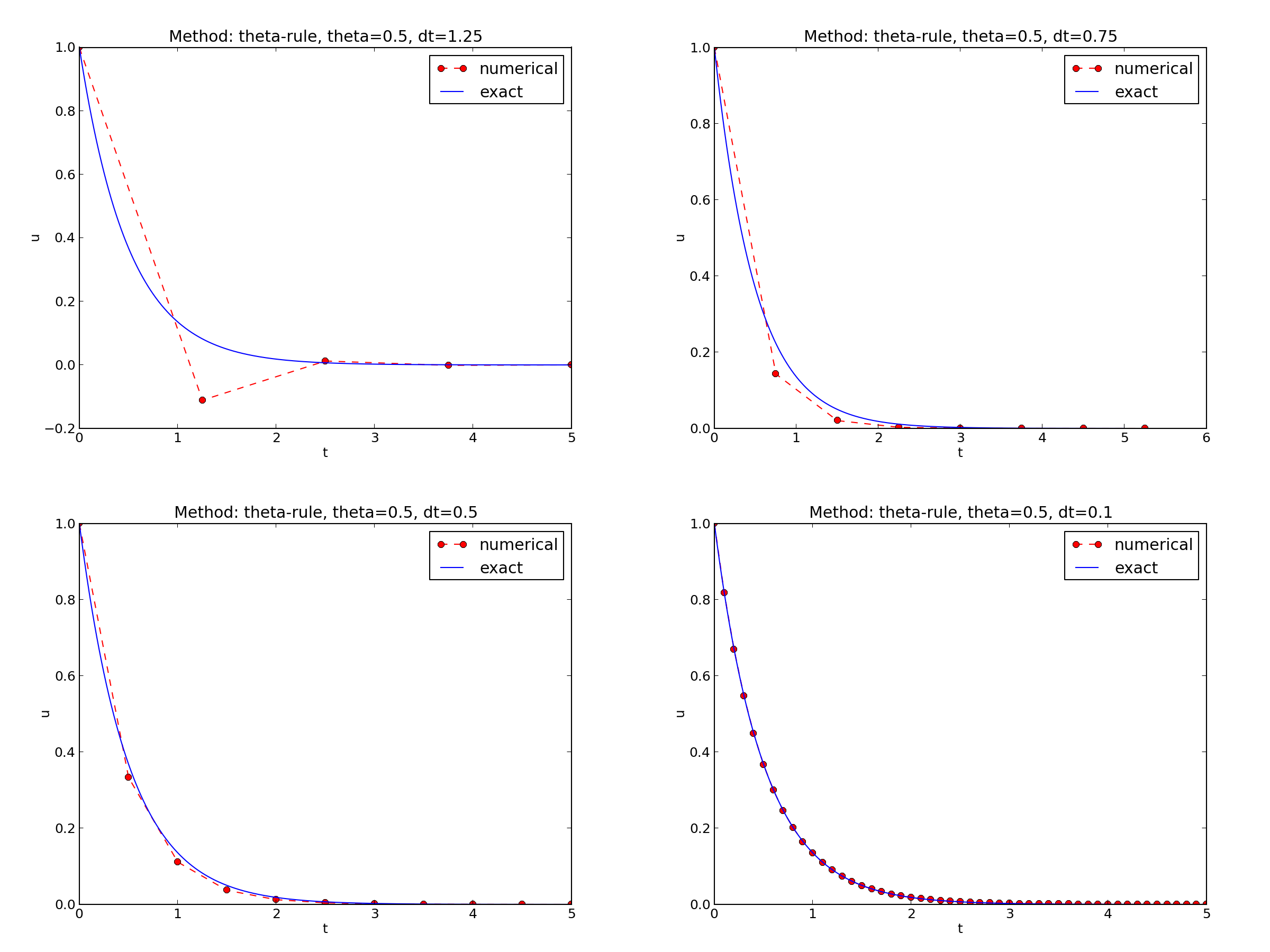
\( I=1 \), \( a=2 \), \( \theta =1,0.5, 0 \), \( \Delta t=1.25, 0.75, 0.5, 0.1 \).
Discouraging numerical solutions; Crank-Nicolson
Discouraging numerical solutions; Forward Euler

Summary of observations
The characteristics of the displayed curves can be summarized as follows:
- The Backward Euler scheme always gives a monotone solution, lying above the exact curve.
- The Crank-Nicolson scheme gives the most accurate results, but for \( \Delta t=1.25 \) the solution oscillates.
- The Forward Euler scheme gives a growing, oscillating solution for \( \Delta t=1.25 \); a decaying, oscillating solution for \( \Delta t=0.75 \); a strange solution \( u^n=0 \) for \( n\geq 1 \) when \( \Delta t=0.5 \); and a solution seemingly as accurate as the one by the Backward Euler scheme for \( \Delta t = 0.1 \), but the curve lies below the exact solution.
Problem setting
- Under what circumstances, i.e., values of the input data \( I \), \( a \), and \( \Delta t \) will the Forward Euler and Crank-Nicolson schemes result in undesired oscillatory solutions?
Techniques of investigation:
- Numerical experiments
- Mathematical analysis
Another question to be raised is
- How does \( \Delta t \) impact the error in the numerical solution?
Experimental investigation of oscillatory solutions
The solution is oscillatory if
$$ u^{n} > u^{n-1},$$
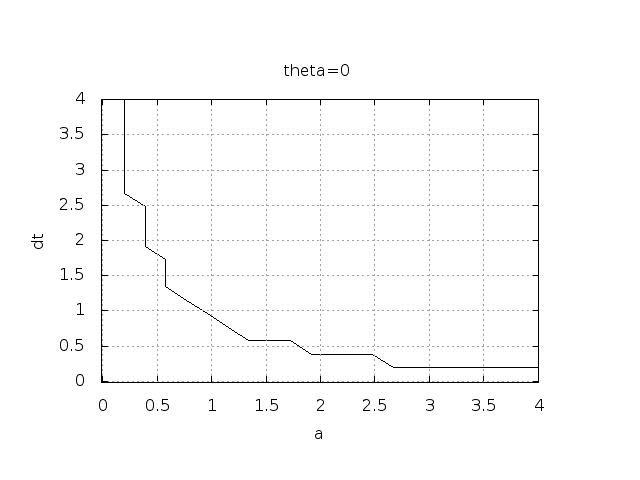
Seems that \( a\Delta t < 1 \) for FE and 2 for CN.
Exact numerical solution
Starting with \( u^0=I \), the simple recursion (30) can be applied repeatedly \( n \) times, with the result that
$$
\begin{equation}
u^{n} = IA^n,\quad A = \frac{1 - (1-\theta) a\Delta t}{1 + \theta a\Delta t}\tp
\tag{31}
\end{equation}
$$
Such an exact discrete solution is unusual, but very handy for analysis.
Stability
Since \( u^n\sim A^n \),
- \( A<0 \) gives a factor \( (-1)^n \) and oscillatory solutions
- \( |A|>1 \) gives growing solutions
- Recall: the exact solution is monotone and decaying
- If these qualitative properties are not met, we say that the numerical solution is unstable
Computation of stability in this problem
\( A<0 \) if
$$
\frac{1 - (1-\theta) a\Delta t}{1 + \theta a\Delta t} < 0
$$
To avoid oscillatory solutions we must have \( A> 0 \) and
$$
\begin{equation}
\Delta t < \frac{1}{(1-\theta)a}\tp
\end{equation}
$$
- Always fulfilled for Backward Euler
- \( \Delta t \leq 1/a \) for Forward Euler
- \( \Delta t \leq 2/a \) for Crank-Nicolson
Computation of stability in this problem
\( |A|\leq 1 \) means \( -1\leq A\leq 1 \)
$$
\begin{equation}
-1\leq\frac{1 - (1-\theta) a\Delta t}{1 + \theta a\Delta t} \leq 1\tp
\tag{32}
\end{equation}
$$
\( -1 \) is the critical limit:
$$
\begin{align*}
\Delta t &\leq \frac{2}{(1-2\theta)a},\quad \theta < \half\\
\Delta t &\geq \frac{2}{(1-2\theta)a},\quad \theta > {\half}
\end{align*}
$$
- Always fulfilled for Backward Euler and Crank-Nicolson
- \( \Delta t \leq 2/a \) for Forward Euler
Explanation of problems with Forward Euler

- \( a\Delta t= 2\cdot 1.25=2.5 \) and \( A=-1.5 \): oscillations and growth
- \( a\Delta t = 2\cdot 0.75=1.5 \) and \( A=-0.5 \): oscillations and decay
- \( \Delta t=0.5 \) and \( A=0 \): \( u^n=0 \) for \( n>0 \)
- Smaller \( Delta t \): qualitatively correct solution
Explanation of problems with Crank-Nicolson

- \( \Delta t=1.25 \) and \( A=-0.25 \): oscillatory solution
- Never any growing solution
Summary of stability
- Forward Euler is conditionally stable
- \( \Delta t < 2/a \) for avoiding growth
- \( \Delta t\leq 1/a \) for avoiding oscillations
- The Crank-Nicolson is unconditionally stable wrt growth and conditionally stable wrt oscillations
- \( \Delta t < 2/a \) for avoiding oscillations
- Backward Euler is unconditionally stable
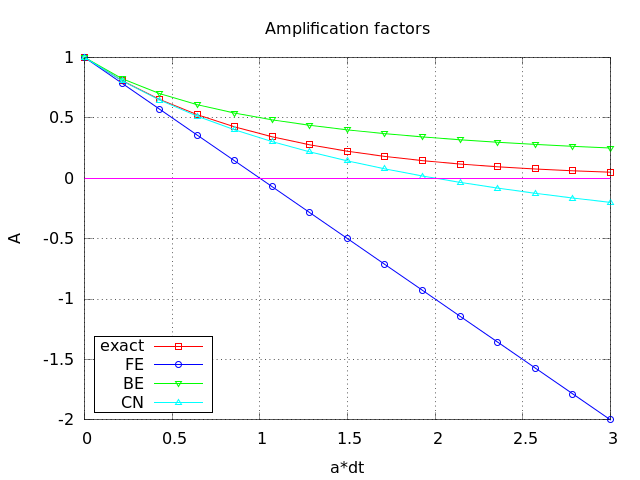
Comparing amplification factors
\( u^{n+1} \) is an amplification \( A \) of \( u^n \):
$$ u^{n+1} = Au^n,\quad A = \frac{1 - (1-\theta) a\Delta t}{1 + \theta a\Delta t} $$
The exact solution is also an amplification:
$$ u(t_{n+1}) = \Aex u(t_n), \quad \Aex = e^{-a\Delta t}$$
A possible measure of accuracy: \( \Aex - A \)
Plot of amplification factors
Series expansion of amplification factors
To investigate \( \Aex - A \) mathematically, we can Taylor expand the expression, using \( p=a\Delta t \) as variable.
>>> from sympy import *
>>> # Create p as a mathematical symbol with name 'p'
>>> p = Symbol('p')
>>> # Create a mathematical expression with p
>>> A_e = exp(-p)
>>>
>>> # Find the first 6 terms of the Taylor series of A_e
>>> A_e.series(p, 0, 6)
1 + (1/2)*p**2 - p - 1/6*p**3 - 1/120*p**5 + (1/24)*p**4 + O(p**6)
>>> theta = Symbol('theta')
>>> A = (1-(1-theta)*p)/(1+theta*p)
>>> FE = A_e.series(p, 0, 4) - A.subs(theta, 0).series(p, 0, 4)
>>> BE = A_e.series(p, 0, 4) - A.subs(theta, 1).series(p, 0, 4)
>>> half = Rational(1,2) # exact fraction 1/2
>>> CN = A_e.series(p, 0, 4) - A.subs(theta, half).series(p, 0, 4)
>>> FE
(1/2)*p**2 - 1/6*p**3 + O(p**4)
>>> BE
-1/2*p**2 + (5/6)*p**3 + O(p**4)
>>> CN
(1/12)*p**3 + O(p**4)
Error in amplification factors
Focus: the error measure \( A-\Aex \) as function of \( \Delta t \) (recall that \( p=a\Delta t \)):
$$
\begin{equation}
A-\Aex = \left\lbrace\begin{array}{ll}
\Oof{\Delta t^2}, & \hbox{Forward and Backward Euler},\\
\Oof{\Delta t^3}, & \hbox{Crank-Nicolson}
\end{array}\right.
\end{equation}
$$
The fraction of numerical and exact amplification factors
Focus: the error measure \( 1-A/\Aex \) as function of \( p=a\Delta t \):
>>> FE = 1 - (A.subs(theta, 0)/A_e).series(p, 0, 4)
>>> BE = 1 - (A.subs(theta, 1)/A_e).series(p, 0, 4)
>>> CN = 1 - (A.subs(theta, half)/A_e).series(p, 0, 4)
>>> FE
(1/2)*p**2 + (1/3)*p**3 + O(p**4)
>>> BE
-1/2*p**2 + (1/3)*p**3 + O(p**4)
>>> CN
(1/12)*p**3 + O(p**4)
Same leading-order terms as for the error measure \( A-\Aex \).
The true/global error at a point
- The error in \( A \) reflects the local error when going from one time step to the next
- What is the global (true) error at \( t_n \)? \( e^n = \uex(t_n) - u^n = Ie^{-at_n} - IA^n \)
- Taylor series expansions of \( e^n \) simplify the expression
Computing the global error at a point
>>> n = Symbol('n')
>>> u_e = exp(-p*n) # I=1
>>> u_n = A**n # I=1
>>> FE = u_e.series(p, 0, 4) - u_n.subs(theta, 0).series(p, 0, 4)
>>> BE = u_e.series(p, 0, 4) - u_n.subs(theta, 1).series(p, 0, 4)
>>> CN = u_e.series(p, 0, 4) - u_n.subs(theta, half).series(p, 0, 4)
>>> FE
(1/2)*n*p**2 - 1/2*n**2*p**3 + (1/3)*n*p**3 + O(p**4)
>>> BE
(1/2)*n**2*p**3 - 1/2*n*p**2 + (1/3)*n*p**3 + O(p**4)
>>> CN
(1/12)*n*p**3 + O(p**4)
Substitute \( n \) by \( t/\Delta t \):
- Forward and Backward Euler: leading order term \( \half ta^2\Delta t \)
- Crank-Nicolson: leading order term \( \frac{1}{12}ta^3\Delta t^2 \)
Convergence
The numerical scheme is convergent if the global error \( e^n\rightarrow 0 \) as \( \Delta t\rightarrow 0 \). If the error has a leading order term \( \Delta t^r \), the convergence rate is of order \( r \).
Integrated errors
Focus: norm of the numerical error
$$ ||e^n||_{\ell^2} = \sqrt{\Delta t\sum_{n=0}^{N_t} ({\uex}(t_n) - u^n)^2}
\tp $$
Forward and Backward Euler:
$$ ||e^n||_{\ell^2} = \frac{1}{4}\sqrt{\frac{T^3}{3}} a^2\Delta t\tp$$
Crank-Nicolson:
$$ ||e^n||_{\ell^2} = \frac{1}{12}\sqrt{\frac{T^3}{3}}a^3\Delta t^2\tp$$
- 1st order for Forward and Backward Euler
- 2nd order for Crank-Nicolson
Truncation error
- How good is the discrete equation?
- Possible answer: see how well \( \uex \) fits the discrete equation
$$ \lbrack D_t u = -au\rbrack^n,$$
i.e.,
$$ \frac{u^{n+1}-u^n}{\Delta t} = -au^n\tp$$
Insert \( \uex \) (which does not in general fulfill this equation):
$$
\begin{equation}
\frac{\uex(t_{n+1})-\uex(t_n)}{\Delta t} + a\uex(t_n) = R^n \neq 0
\tp
\tag{33}
\end{equation}
$$
Computation of the truncation error
- The residual \( R^n \) is the truncation error.
- How does \( R^n \) vary with \( \Delta t \)?
Tool: Taylor expand \( \uex \) around the point where the ODE is sampled (here \( t_n \))
$$ \uex(t_{n+1}) = \uex(t_n) + \uex'(t_n)\Delta t + \half\uex''(t_n)
\Delta t^2 + \cdots $$
Inserting this Taylor series in (33) gives
$$ R^n = \uex'(t_n) + \half\uex''(t_n)\Delta t + \ldots + a\uex(t_n)\tp$$
Now, \( \uex \) solves the ODE \( \uex'=-a\uex \), and then
$$ R^n \approx \half\uex''(t_n)\Delta t \tp $$
This is a mathematical expression for the truncation error.
The truncation error for other schemes
Backward Euler:
$$ R^n \approx -\half\uex''(t_n)\Delta t, $$
Crank-Nicolson:
$$ R^{n+\half} \approx \frac{1}{24}\uex'''(t_{n+\half})\Delta t^2\tp$$
Consistency, stability, and convergence
- Truncation error measures the residual in the difference equations. The scheme is consistent if the truncation error goes to 0 as \( \Delta t\rightarrow 0 \). Importance: the difference equations approaches the differential equation as \( \Delta t\rightarrow 0 \).
- Stability means that the numerical solution exhibits the same qualitative properties as the exact solution. Here: monotone, decaying function.
- Convergence implies that the true (global) error \( e^n =\uex(t_n)-u^n\rightarrow 0 \) as \( \Delta t\rightarrow 0 \). This is really what we want!
The Lax equivalence theorem for linear differential equations: consistency + stability is equivalent with convergence.
(Consistency and stability is in most problems much easier to establish than convergence.)
Model extensions
Extension to a variable coefficient; Forward and Backward Euler
$$
\begin{equation}
u'(t) = -a(t)u(t),\quad t\in (0,T],\quad u(0)=I \tp
\tag{34}
\end{equation}
$$
The Forward Euler scheme:
$$
\begin{equation}
\frac{u^{n+1} - u^n}{\Delta t} = -a(t_n)u^n
\tp
\end{equation}
$$
The Backward Euler scheme:
$$
\begin{equation}
\frac{u^{n} - u^{n-1}}{\Delta t} = -a(t_n)u^n
\tp
\end{equation}
$$
Extension to a variable coefficient; Crank-Nicolson
Eevaluting \( a(t_{n+\half}) \) and using an average for \( u \):
$$
\begin{equation}
\frac{u^{n+1} - u^{n}}{\Delta t} = -a(t_{n+\half})\half(u^n + u^{n+1})
\tp
\end{equation}
$$
Using an average for \( a \) and \( u \):
$$
\begin{equation}
\frac{u^{n+1} - u^{n}}{\Delta t} = -\half(a(t_n)u^n + a(t_{n+1})u^{n+1})
\tp
\end{equation}
$$
Extension to a variable coefficient; \( \theta \)-rule
The \( \theta \)-rule unifies the three mentioned schemes,
$$
\begin{equation}
\frac{u^{n+1} - u^{n}}{\Delta t} = -a((1-\theta)t_n + \theta t_{n+1})((1-\theta) u^n + \theta u^{n+1})
\tp
\end{equation}
$$
or,
$$
\begin{equation}
\frac{u^{n+1} - u^{n}}{\Delta t} = -(1-\theta) a(t_n)u^n - \theta
a(t_{n+1})u^{n+1}
\tp
\end{equation}
$$
Extension to a variable coefficient; operator notation
$$
\begin{align*}
\lbrack D^+_t u &= -au\rbrack^n,\\
\lbrack D^-_t u &= -au\rbrack^n,\\
\lbrack D_t u &= -a\overline{u}^t\rbrack^{n+\half},\\
\lbrack D_t u &= -\overline{au}^t\rbrack^{n+\half},\\
\tp
\end{align*}
$$
Extension to a source term
$$
\begin{equation}
u'(t) = -a(t)u(t) + b(t),\quad t\in (0,T],\quad u(0)=I
\tp
\tag{35}
\end{equation}
$$
$$
\begin{align*}
\lbrack D^+_t u &= -au + b\rbrack^n,\\
\lbrack D^-_t u &= -au + b\rbrack^n,\\
\lbrack D_t u &= -a\overline{u}^t + b\rbrack^{n+\half},\\
\lbrack D_t u &= \overline{-au+b}^t\rbrack^{n+\half}
\tp
\end{align*}
$$
Implementation of the generalized model problem
$$
\begin{equation}
u^{n+1} = ((1 - \Delta t(1-\theta)a^n)u^n
+ \Delta t(\theta b^{n+1} + (1-\theta)b^n))(1 + \Delta t\theta a^{n+1})^{-1}
\tp
\end{equation}
$$
Implementation where \( a(t) \) and \( b(t) \) are given as Python functions (see file decay_vc.py):
def solver(I, a, b, T, dt, theta):
"""
Solve u'=-a(t)*u + b(t), u(0)=I,
for t in (0,T] with steps of dt.
a and b are Python functions of t.
"""
dt = float(dt) # avoid integer division
Nt = int(round(T/dt)) # no of time intervals
T = Nt*dt # adjust T to fit time step dt
u = zeros(Nt+1) # array of u[n] values
t = linspace(0, T, Nt+1) # time mesh
u[0] = I # assign initial condition
for n in range(0, Nt): # n=0,1,...,Nt-1
u[n+1] = ((1 - dt*(1-theta)*a(t[n]))*u[n] + \
dt*(theta*b(t[n+1]) + (1-theta)*b(t[n])))/\
(1 + dt*theta*a(t[n+1]))
return u, t
Implementations of variable coefficients; functions
Plain functions:
def a(t):
return a_0 if t < tp else k*a_0
def b(t):
return 1
Implementations of variable coefficients; classes
Better implementation: class with the parameters a0, tp, and k
as attributes and a special method __call__ for evaluating \( a(t) \):
class A:
def __init__(self, a0=1, k=2):
self.a0, self.k = a0, k
def __call__(self, t):
return self.a0 if t < self.tp else self.k*self.a0
a = A(a0=2, k=1) # a behaves as a function a(t)
Implementations of variable coefficients; lambda function
Quick writing: a one-liner lambda function
a = lambda t: a_0 if t < tp else k*a_0
In general,
f = lambda arg1, arg2, ...: expressin
is equivalent to
def f(arg1, arg2, ...):
return expression
One can use lambda functions directly in calls:
u, t = solver(1, lambda t: 1, lambda t: 1, T, dt, theta)
for a problem \( u'=-u+1 \), \( u(0)=1 \).
A lambda function can appear anywhere where a variable can appear.
Verification via trivial solutions
- Start debugging of a new code with trying a problem where \( u=\hbox{const} \neq 0 \).
- Choose \( u=C \) (a constant). Choose any \( a(t) \) and set \( b=a(t)C \) and \( I=C \).
- "All" numerical methods will reproduce \( u=_{\hbox{const}} \) exactly (machine precision).
- Often \( u=C \) eases debugging.
- In this example: any error in the formula for \( u^{n+1} \) make \( u\neq C \)!
Verification via trivial solutions; nose test
import nose.tools as nt
def test_constant_solution():
"""
Test problem where u=u_const is the exact solution, to be
reproduced (to machine precision) by any relevant method.
"""
def exact_solution(t):
return u_const
def a(t):
return 2.5*(1+t**3) # can be arbitrary
def b(t):
return a(t)*u_const
u_const = 2.15
theta = 0.4; I = u_const; dt = 4
Nt = 4 # enough with a few steps
u, t = solver(I=I, a=a, b=b, T=Nt*dt, dt=dt, theta=theta)
print u
u_e = exact_solution(t)
difference = abs(u_e - u).max() # max deviation
nt.assert_almost_equal(difference, 0, places=14)
Verification via manufactured solutions
- Choose any formula for \( u(t) \).
- Fit \( I \), \( a(t) \), and \( b(t) \) in \( u'=-au+b \), \( u(0)=I \), to make the chosen formula a solution of the ODE problem.
- Then we can always have an analytical solution (!).
- Ideal for verification: testing convergence rates.
- Called the method of manufactured solutions (MMS)
- Special case: \( u \) linear in \( t \), because all sound numerical methods will reproduce a linear \( u \) exactly (machine precision).
- \( u(t) = ct + d \). \( u(0)=0 \) means \( d=I \).
- ODE implies \( c = -a(t)u + b(t) \).
- Choose \( a(t) \) and \( c \), and set \( b(t) = c + a(t)(ct + I) \).
- Any error in the formula for \( u^{n+1} \) makes \( u\neq ct+I \)!
Linear manufactured solution
\( u^n = ct_n+I \) fulfills the discrete equations!
First,
$$
\begin{align}
\lbrack D_t^+ t\rbrack^n &= \frac{t_{n+1}-t_n}{\Delta t}=1,
\tag{36}\\
\lbrack D_t^- t\rbrack^n &= \frac{t_{n}-t_{n-1}}{\Delta t}=1,
\tag{37}\\
\lbrack D_t t\rbrack^n &= \frac{t_{n+\half}-t_{n-\half}}{\Delta t}=\frac{(n+\half)\Delta t - (n-\half)\Delta t}{\Delta t}=1\tag{38}
\tp
\end{align}
$$
Forward Euler:
$$ [D^+ u = -au + b]^n, $$
\( a^n=a(t_n) \), \( b^n=c + a(t_n)(ct_n + I) \), and \( u^n=ct_n + I \) results in
$$ c = -a(t_n)(ct_n+I) + c + a(t_n)(ct_n + I) = c $$
Nose test for linear manufactured solution
def test_linear_solution():
"""
Test problem where u=c*t+I is the exact solution, to be
reproduced (to machine precision) by any relevant method.
"""
def exact_solution(t):
return c*t + I
def a(t):
return t**0.5 # can be arbitrary
def b(t):
return c + a(t)*exact_solution(t)
theta = 0.4; I = 0.1; dt = 0.1; c = -0.5
T = 4
Nt = int(T/dt) # no of steps
u, t = solver(I=I, a=a, b=b, T=Nt*dt, dt=dt, theta=theta)
u_e = exact_solution(t)
difference = abs(u_e - u).max() # max deviation
print difference
# No of decimal places for comparison depend on size of c
nt.assert_almost_equal(difference, 0, places=14)
Extension to systems of ODEs
Sample system:
$$
\begin{align}
u' &= a u + bv,\\
v' &= cu + dv,
\end{align}
$$
The Forward Euler method:
$$
\begin{align}
u^{n+1} &= u^n + \Delta t (a u^n + b v^n),\\
v^{n+1} &= u^n + \Delta t (cu^n + dv^n)
\tp
\end{align}
$$
The Backward Euler method gives a system of algebraic equations
The Backward Euler scheme:
$$
\begin{align}
u^{n+1} &= u^n + \Delta t (a u^{n+1} + b v^{n+1}),\\
v^{n+1} &= v^n + \Delta t (c u^{n+1} + d v^{n+1})\tp
\end{align}
$$
which is a \( 2\times 2 \) linear system:
$$
\begin{align}
(1 - \Delta t a)u^{n+1} + bv^{n+1} &= u^n ,\\
c u^{n+1} + (1 - \Delta t d) v^{n+1} &= v^n ,
\end{align}
$$
Crank-Nicolson also gives a \( 2\times 2 \) linear system.
General first-order ODEs
Generic form
The standard form for ODEs:
$$
\begin{equation}
u' = f(u,t),\quad u(0)=I,
\tag{39}
\end{equation}
$$
\( u \) and \( f \): scalar or vector.
Vectors in case of ODE systems:
$$ u(t) = (u^{(0)}(t),u^{(1)}(t),\ldots,u^{(m-1)}(t)) \tp $$
$$
\begin{align*}
f(u, t) = ( & f^{(0)}(u^{(0)},\ldots,u^{(m-1)}),\\
& f^{(1)}(u^{(0)},\ldots,u^{(m-1)}),\\
& \vdots\\
& f^{(m-1)}(u^{(0)}(t),\ldots,u^{(m-1)}(t)))
\tp
\end{align*}
$$
The \( \theta \)-rule
$$
\begin{equation}
\frac{u^{n+1}-u^n}{\Delta t} = \theta f(u^{n+1},t_{n+1}) +
(1-\theta)f(u^n, t_n)\tp
\tag{40}
\end{equation}
$$
Bringing the unknown \( u^{n+1} \) to the left-hand side and the known terms
on the right-hand side gives
$$
\begin{equation}
u^{n+1} - \Delta t \theta f(u^{n+1},t_{n+1}) =
u^n + \Delta t(1-\theta)f(u^n, t_n)\tp
\end{equation}
$$
This is a nonlinear equation in \( u^{n+1} \) (unless \( f \) is linear in \( u \))!
Implicit 2-step backward scheme
$$ u'(t_{n+1}) \approx \frac{3u^{n+1} - 4u^{n} + u^{n-1}}{2\Delta t},$$
Scheme:
$$ u^{n+1} = \frac{4}{3}u^n - \frac{1}{3}u^{n-1} +
\frac{2}{3}\Delta t f(u^{n+1}, t_{n+1})
\tp
\tag{41}
$$
Nonlinear equation for \( u^{n+1} \).
The Leapfrog scheme
Idea:
$$
\begin{equation}
u'(t_n)\approx \frac{u^{n+1}-u^{n-1}}{2\Delta t} = [D_{2t} u]^n,
\end{equation}
$$
Scheme:
$$ [D_{2t} u = f(u,t)]^n,$$
or written out,
$$
\begin{equation}
u^{n+1} = u^{n-1} + \Delta t f(u^n, t_n)
\tp
\tag{42}
\end{equation}
$$
- Some other scheme must be used as starter (\( u^1 \)).
- Explicit scheme - a nonlinear \( f \) (in \( u \)) is trivial to handle.
- Downside: Leapfrog is always unstable after some time.
The filtered Leapfrog scheme
After computing \( u^{n+1} \), stabilize Leapfrog by
$$
\begin{equation}
u^n\ \leftarrow\ u^n + \gamma (u^{n-1} - 2u^n + u^{n+1})
\tag{43}
\tp
\end{equation}
$$
2nd-order Runge-Kutta scheme
Forward-Euler + approximate Crank-Nicolson:
$$
\begin{align}
u^* &= u^n + \Delta t f(u^n, t_n),
\tag{44}\\
u^{n+1} &= u^n + \Delta t \half \left( f(u^n, t_n) + f(u^*, t_{n+1})
\right),
\tag{45}
\end{align}
$$
4th-order Runge-Kutta scheme
- The most famous and widely used ODE method
- 4 evaluations of \( f \) per time step
- Its derivation is a very good illustration of numerical thinking!
2nd-order Adams-Bashforth scheme
$$
\begin{equation}
u^{n+1} = u^n + \half\Delta t\left( 3f(u^n, t_n) - f(u^{n-1}, t_{n-1})
\right)
\tp
\tag{46}
\end{equation}
$$
3rd-order Adams-Bashforth scheme
$$
\begin{equation}
u^{n+1} = u^n + \frac{1}{12}\left( 23f(u^n, t_n) - 16 f(u^{n-1},t_{n-1})
+ 5f(u^{n-2}, t_{n-2})\right)
\tp
\tag{47}
\end{equation}
$$
The Odespy software
Odespy features simple Python implementations of the most fundamental schemes as well as Python interfaces to several famous packages for solving ODEs: ODEPACK, Vode, rkc.f, rkf45.f, Radau5, as well as the ODE solvers in SciPy, SymPy, and odelab.
Typical usage:
# Define right-hand side of ODE
def f(u, t):
return -a*u
import odespy
import numpy as np
# Set parameters and time mesh
I = 1; a = 2; T = 6; dt = 1.0
Nt = int(round(T/dt))
t_mesh = np.linspace(0, T, Nt+1)
# Use a 4th-order Runge-Kutta method
solver = odespy.RK4(f)
solver.set_initial_condition(I)
u, t = solver.solve(t_mesh)
Example: Runge-Kutta methods
solvers = [odespy.RK2(f),
odespy.RK3(f),
odespy.RK4(f),
odespy.BackwardEuler(f, nonlinear_solver='Newton')]
for solver in solvers:
solver.set_initial_condition(I)
u, t = solver.solve(t)
# + lots of plot code...
Plots from the experiments
The 4-th order Runge-Kutta method (RK4) is the method of choice!
Example: Adaptive Runge-Kutta methods
- Adaptive methods find "optimal" locations of the mesh points to ensure that the error is less than a given tolerance.
- Downside: approximate error estimation, not always optimal location of points.
- "Industry standard ODE solver": Dormand-Prince 4/5-th order
Runge-Kutta (MATLAB's famous
ode45).