Pure Python code for Monte Carlo simulation
The computational problem
A scalar Python implementation
A vectorized Python implementation
Migrating scalar Python code to Cython
A plain Cython implementation
A better Cython implementation
Migrating code to C
Writing a C program
Migrating loops to C code via F2PY
Migrating loops to C code via Cython
Comparing efficiency
References
Monte Carlo simulations are usually known to require long execution times. Implementing such simulations in pure Python may lead to inefficient code. The purpose of this note is to show how Python implementations of Monte Carlo simulations, can be made much more efficient by porting the code to Cython. Pure C implementations are included for comparison of efficiency. The reader should know about basic Python and perhaps a bit about Monte Carlo simulations. Cython will be introduced in a step-by-step fashion
A short, intuitive algorithm in Python is first developed. Then this code is vectorized using functionality of the Numerical Python package. Later sections migrate the algorithm to Cython code and also plain C code for comparison. At the end the various techniques are ranked according to their computational efficiency.
A die is thrown \( m \) times. What is the probability of getting six eyes at least \( n \) times? For example, if \( m=5 \) and \( n=3 \), this is the same as asking for the probability that three or more out of five dice show six eyes.
The probability can be estimated by Monte Carlo simulation. We simulate the process a large number of times, \( N \), and count how many times, \( M \), the experiment turned out successfully, i.e., when we got at least \( n \) out of \( m \) dice with six eyes in a throw.
Monte Carlo simulation has traditionally been viewed as a very costly computational method, normally requiring very sophisticated, fast computer implementations in compiled languages. An interesting question is how useful high-level languages like Python and associated tools are for Monte Carlo simulation. This will now be explored.
Let us introduce the more descriptive variables ndice for \( m \)
and nsix for \( n \). The Monte Carlo method is
simply a loop, repeated N times, where the body of the loop may
directly express the problem at hand. Here, we draw ndice random
integers r in \( [1,6] \) inside the loop and count of many (six) that
equal 6. If six >= nsix, the experiment is a success and we increase
the counter M by one.
A Python function implementing this approach may look as follows:
import random
def dice6_py(N, ndice, nsix):
M = 0 # no of successful events
for i in range(N): # repeat N experiments
six = 0 # how many dice with six eyes?
for j in range(ndice):
r = random.randint(1, 6) # roll die no. j
if r == 6:
six += 1
if six >= nsix: # successful event?
M += 1
p = float(M)/N
return p
The float(M) transformation is important since M/N will imply integer
division when M and N both are integers in Python v2.x and many other
languages.
We will refer to this implementation is the plain Python implementation. Timing the function can be done by:
import time
t0 = time.clock()
p = dice6_py(N, ndice, nsix)
t1 = time.clock()
print 'CPU time for loops in Python:', t1-t0
The function above can be verified by studying the (somewhat simplified) case \( m=n \) where the probability becomes \( 6^{-n} \). The probability quickly becomes small with increasing \( n \). For such small probabilities the number of successful events \( M \) is small, and \( M/N \) will not be a good approximation to the probability unless \( M \) is reasonably large, which requires a very large \( N \). For example, with \( n=4 \) and \( N=10^5 \) the average probability in 25 full Monte Carlo experiments is 0.00078 while the exact answer is 0.00077. With \( N=10^6 \) we get the two correct significant digits from the Monte Carlo simulation, but the extra digit costs a factor of 10 in computing resources since the CPU time scales linearly with \( N \).
A vectorized version of the previous program consists of replacing the
explicit loops in Python by efficient operations on vectors or arrays,
using functionality in the Numerical Python (numpy) package.
Each array operation takes place in C or Fortran and is hence
much more efficient than the corresponding loop version in Python.
First, we must generate all the random numbers to be used in one
operation, which runs fast since all numbers are then calculated in
efficient C code. This is accomplished using the numpy.random
module. Second, the analysis of the large collection of random
numbers must be done by appropriate vector/array operations such that
no looping in Python is needed. The solution algorithm must therefore
be expressed through a series of function calls to the numpy library.
Vectorization requires knowledge of the library's functionality and
how to assemble the relevant building blocks to an algorithm without
operations on individual array elements.
Generation of ndice random number of eyes for N experiments is
performed by
import numpy as np
eyes = np.random.random_integers(1, 6, size=(N, ndice))
eyes array corresponds to one Monte Carlo experiment.
The next step is to count the number of successes in each experiment.
This counting should not make use of any loop. Instead we can test
eyes == 6 to get a boolean array where an element i,j is True if
throw (or die) number j in Monte Carlo experiment number i gave
six eyes. Summing up the rows in this boolean array (True is interpreted
as 1 and False as 0), we are
interested in the rows where the sum is equal to or greater than
nsix, because the number of such rows equals the number of
successful events. The vectorized algorithm can be expressed as
def dice6_vec1(N, ndice, nsix):
eyes = np.random.random_integers(1, 6, size=(N, ndice))
compare = eyes == 6
throws_with_6 = np.sum(compare, axis=1) # sum over columns
nsuccesses = throws_with_6 >= nsix
M = np.sum(nsuccesses)
p = float(M)/N
return p
The use of np.sum instead of Python's own sum function is
essential for the speed of this function: using M = sum(nsucccesses)
instead slows down the code by a factor of almost 10! We shall refer
to the dice6_vec1 function as the vectorized Python, version1
implementation.
The criticism against the vectorized version is that the original
problem description, which was almost literally turned into Python
code in the dice6_py function, has now become much more
complicated. We have to decode the calls to various
numpy functionality to actually realize that dice6_py
and dice6_vec correspond to the same mathematics.
Here is another possible vectorized algorithm, which is easier to understand, because we retain the Monte Carlo loop and vectorize only each individual experiment:
def dice6_vec2(N, ndice, nsix):
eyes = np.random.random_integers(1, 6, (N, ndice))
six = [6 for i in range(ndice)]
M = 0
for i in range(N):
# Check experiment no. i:
compare = eyes[i,:] == six
if np.sum(compare) >= nsix:
M += 1
p = float(M)/N
return p
We refer to this implementation as vectorized Python, version 2. As will be shown later, this implementation is significantly slower than the plain Python implementation (!) and very much slower than the vectorized Python, version 1 approach. A conclusion is that readable, partially vectorized code, may run slower than straightforward scalar code.
A Cython program starts with the scalar Python implementation, but all
variables are specified with their types, using Cython's variable
declaration syntax, like cdef int M = 0 where we in standard Python
just write M = 0. Adding such variable declarations in the scalar
Python implementation is straightforward:
import random
def dice6_cy1(int N, int ndice, int nsix):
cdef int M = 0 # no of successful events
cdef int six, r
cdef double p
for i in range(N): # repeat N experiments
six = 0 # how many dice with six eyes?
for j in range(ndice):
r = random.randint(1, 6) # roll die no. j
if r == 6:
six += 1
if six >= nsix: # successful event?
M += 1
p = float(M)/N
return p
This code must be put in a separate file with extension .pyx.
Running Cython on this file translates the Cython code to C.
Thereafter, the C code must be compiled and linked to form a
shared library, which can be imported in Python as a module. All
these tasks are normally automated by a setup.py script. Let the
dice6_cy1 function above be stored in a file dice6.pyx. A proper
setup.py script looks as follows:
from distutils.core import setup
from distutils.extension import Extension
from Cython.Distutils import build_ext
setup(
name='Monte Carlo simulation',
ext_modules=[Extension('_dice6_cy', ['dice6.pyx'],)],
cmdclass={'build_ext': build_ext},
)
Running
Terminal> python setup.py build_ext --inplace
_dice6_cy.so (known as a C extension module) which
can be loaded into Python as a module with name _dice6_cy:
from _dice6_cy import dice6_cy1
import time
t0 = time.clock()
p = dice6_cy1(N, ndice, nsix)
t1 = time.clock()
print t1 - t0
dice6_cy1 function
are turned into plain and fast C code,
the speed is not much improved compared with the original scalar
Python code.
To investigate what takes time in this Cython implementation, we can perform
a profiling. The template for profiling a Python function whose
call syntax is stored in some string statement, reads
import cProfile, pstats
cProfile.runctx(statement, globals(), locals(), 'tmp_profile.dat')
s = pstats.Stats('tmp_profile.dat')
s.strip_dirs().sort_stats('time').print_stats(30)
tmp_profile.dat.
Our interest now is the dice6_cy1 function so we set
statement = 'dice6_cy1(N, ndice, nsix)'
# cython: profile=True
5400004 function calls in 7.525 CPU seconds
Ordered by: internal time
ncalls tottime percall cumtime percall filename:lineno(function)
1800000 4.511 0.000 4.863 0.000 random.py:160(randrange)
1800000 1.525 0.000 6.388 0.000 random.py:224(randint)
1 1.137 1.137 7.525 7.525 dice6.pyx:6(dice6_cy1)
1800000 0.352 0.000 0.352 0.000 {method 'random' ...
1 0.000 0.000 7.525 7.525 {dice6_cy.dice6_cy1}
random.randint that consumes
almost all the time. The reason is that the generated C code must
call a Python module (random), which implies a lot of overhead. The C code
should only call plain C functions, or if Python functions must
be called, they should involve so much computations that the overhead
in calling Python from C is negligible.
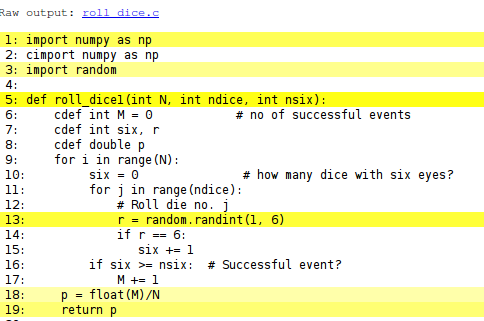
Instead of profiling the code to uncover inefficient constructs we can generate a visual representation of how the Python code is translated to C. Running
Terminal> cython -a dice6.pyx
dice6.html which can be loaded into a web browser
to inspect what Cython has done with the Python code.

White lines indicate that the Python code is translated into C code, while
the yellow lines indicate that the generated C code must make calls
back to Python (using the Python C API, which implies overhead). Here,
the random.randint call is in yellow, so this call is
not translated to efficient C code.
To speed up the previous Cython code, we have to get rid of the
random.randint call every time we need a random variable. Either we
must call some C function for generating a random variable or we must
create a bunch of random numbers simultaneously as we did in the
vectorized functions shown above. We first try the latter well-known
strategy and apply the numpy.random module to generate all the
random numbers we need at once:
import numpy as np
cimport numpy as np
@cython.boundscheck(False) # turn off array bounds check
@cython.wraparound(False) # turn off negative indices ([-1,-1])
def dice6_cy2(int N, int ndice, int nsix):
# Use numpy to generate all random numbers
...
cdef np.ndarray[np.int_t, ndim=2, mode='c'] eyes = \
np.random.random_integers(1, 6, (N, ndice))
cimport statement imports
a special version of numpy for Cython and is needed after the
standard numpy import. The declaration of the array of
random numbers could just go as
cdef np.ndarray eyes = np.random.random_integers(1, 6, (N, ndice))
eyes array will then be slow because
Cython does not have enough information about the array. To generate
optimal C code, we must provide information on the element types in
the array, the number of dimensions of the array, that the array is
stored in contiguous memory, that we do not want the overhead of
checking whether indices are within their bounds or not, and that we
do not need negative indices (which slows down array indexing). The
latter two properties are taken care of by the
@cython.boundscheck(False) and the @cython.wraparound(False)
statements (decorators) right before the function,
respectively, while the rest of
the information is specified within square brackets in the cdef
np.ndarray declaration. Inside the brackets, np.int_t denotes
integer array elements (np.int is the usual data type object, but
np.int_t is a Cython precompiled version of this object), ndim=2
tells that the array has two dimensions (indices),
and mode='c' indicates contiguous
storage of the array. With all this extra information, Cython can generate C
code that works with numpy arrays as efficiently as native C arrays.
The rest of the code is a plain copy of the dice6_py function, but
with the random.randint call replaced by an array look-up
eyes[i,j] to retrieve the next random number. The two loops will
now be as efficient as if they were coded directly in pure C.
The complete code for the efficient version of the dice6_cy1 function
looks as follows:
import numpy as np
cimport numpy as np
import cython
@cython.boundscheck(False) # turn off array bounds check
@cython.wraparound(False) # turn off negative indices ([-1,-1])
def dice6_cy2(int N, int ndice, int nsix):
# Use numpy to generate all random numbers
cdef int M = 0 # no of successful events
cdef int six, r
cdef double p
cdef np.ndarray[np.int_t, ndim=2, mode='c'] eyes = \
np.random.random_integers(1, 6, (N, ndice))
for i in range(N):
six = 0 # how many dice with six eyes?
for j in range(ndice):
r = eyes[i,j] # roll die no. j
if r == 6:
six += 1
if six >= nsix: # successful event?
M += 1
p = float(M)/N
return p
The disadvantage with the dice6_cy2 function is that large simulations
(large N) also require large amounts of memory, which usually limits
the possibility for high accuracy much more than the CPU time. It would
be advantageous to have a fast random number generator a la
random.randint in C. The C library stdlib has a generator of
random integers, rand(), generating numbers from 0 to up RAND_MAX.
Both the rand function and the RAND_MAX integer are easy to
access in a Cython program:
from libc.stdlib cimport rand, RAND_MAX
r = 1 + int(6.0*rand()/RAND_MAX) # random integer 1,...,6
rand() returns an integer so we must avoid integer
division by ensuring that the denominator is a real number. We also
need to explicitly convert the resulting real fraction to int
since r is declared as int.
With this way of generating random numbers we can create a version of
dice6_cy1 that is as fast as dice6_cy, but avoids all the memory
demands and the somewhat complicated array declarations of the latter:
from libc.stdlib cimport rand, RAND_MAX
def dice6_cy3(int N, int ndice, int nsix):
cdef int M = 0 # no of successful events
cdef int six, r
cdef double p
for i in range(N):
six = 0 # how many dice with six eyes?
for j in range(ndice):
# Roll die no. j
r = 1 + int(6.0*rand()/RAND_MAX)
if r == 6:
six += 1
if six >= nsix: # successful event?
M += 1
p = float(M)/N
return p
A natural next improvement would be to program the Monte Carlo
simulation loops directly in a compiled programming language, which
guarantees optimal speed. Here we choose the C programming
language for this purpose.
The C version of our dice6 function and an associated main program
take the form
#include <stdio.h>
#include <stdlib.h>
double dice6(int N, int ndice, int nsix)
{
int M = 0;
int six, r, i, j;
double p;
for (i = 0; i < N; i++) {
six = 0;
for (j = 0; j < ndice; j++) {
r = 1 + rand()/(RAND_MAX*6.0); /* roll die no. j */
if (r == 6)
six += 1;
}
if (six >= nsix)
M += 1;
}
p = ((double) M)/N;
return p;
}
int main(int nargs, const char* argv[])
{
int N = atoi(argv[1]);
int ndice = 6;
int nsix = 3;
double p = dice6(N, ndice, nsix);
printf("C code: N=%d, p=%.6f\n", N, p);
return 0;
}
This code is placed in a file dice6_c.c.
The file can typically be compiled and run by
Terminal> gcc -O3 -o dice6.capp dice6_c.c
Terminal> ./dice6.capp 1000000
Instead of programming the whole application in C, we may consider
migrating the loops to the C function dice6 shown above and then
have the rest of the program (essentially the calling main program) in
Python. This is a convenient solution if we were to do many other,
less CPU time critical things for convenience in Python.
There are many alternative techniques for calling C functions from
Python. Here we shall explain two. The first applies the program
f2py to generate the necessary code that glues Python and C.
The f2py program was actually made for gluing Python and Fortran,
but it can work with C too. We need a specification of the C
function to call in terms of a Fortran 90 module. Such a module can
be written by hand, but f2py can also generate it. To this end,
we make a Fortran file dice6_c_signature.f
with the signature of the C function written
in Fortran 77 syntax with some annotations:
real*8 function dice6(n, ndice, nsix)
Cf2py intent(c) dice6
integer n, ndice, nsix
Cf2py intent(c) n, ndice, nsix
return
end
The annotations intent(c) are necessary to tell f2py that the
Fortran variables are to be treated as plain C variables and
not as pointers (which is the default interpretation of variables
in Fortran). The C2fpy are special comment lines that f2py
recognizes, and these lines are used to provide extra information
to f2py which have no meaning in plain Fortran 77.
We must run f2py to generate a .pyf file with a Fortran 90
module specification of the C function to call:
Terminal> f2py -m _dice6_c1 -h dice6_c.pyf \
dice6_c_signature.f
_dice6_c1 is the name of the module with the C function
that is to be imported in Python, and dice6_c.pyf is the
name of the Fortran 90 module file to be generated. Programmers
who know Fortran 90 may want to write the dice6_c.pyf file
by hand.
The next step is to use the information in dice6_c.pyf
to generate a (C extension) module _dice6_c1. Fortunately,
f2py generates the necessary code, and compiles and links the relevant
files, to form a shared library file _dice6_c1.so, by a short command:
Terminal> f2py -c dice6_c.pyf dice6_c.c
>>> import _dice6_c1
>>> print dir(_dice6_c1) # module contents
['__doc__', '__file__', '__name__', '__package__',
'__version__', 'dice6']
>>> print _dice6_c1.dice6.__doc__
dice6 - Function signature:
dice6 = dice6(n,ndice,nsix)
Required arguments:
n : input int
ndice : input int
nsix : input int
Return objects:
dice6 : float
>>> _dice6_c1.dice6(N=1000, ndice=4, nsix=2)
0.145
dice6 via an f2py
generated module is referred to as C via f2py.
The Cython tool can also be used to call C code, not only generating
C code from the Cython language.
Our C code is in the file dice6_c.c, but for Cython to
see this code we need to create a header file dice6_c.h
listing the definition of the function(s) we want to call from Python.
The header file takes the form
extern double dice6(int N, int ndice, int nsix);
The next step is to make a .pyx file with a definition of the C
function from the header file and a Python function that calls
the C function:
cdef extern from "dice6_c.h":
double dice6(int N, int ndice, int nsix)
def dice6_cwrap(int N, int ndice, int nsix):
return dice6(N, ndice, nsix)
Cython must use this file, named dice6_cwrap.pyx,
to generate C code, which is to be compiled and linked with
the dice6_c.c code. All this is accomplished in a setup.py
script:
from distutils.core import setup
from distutils.extension import Extension
from Cython.Distutils import build_ext
sources = ['dice6_cwrap.pyx', 'dice6_c.c']
setup(
name='Monte Carlo simulation',
ext_modules=[Extension('_dice6_c2', sources)],
cmdclass={'build_ext': build_ext},
)
setup.py script is run as
Terminal> python setup.py build_ext --inplace
_dice6_c2.so, which can
be loaded into Python as a module:
>>> import _dice6_c2
>>> print dir(_dice6_c2)
['__builtins__', '__doc__', '__file__', '__name__',
'__package__', '__test__', 'dice6_cwrap']
dice6_cwrap, which
was made to call the underlying C function dice6.
All the files corresponding to the various techniques described above
are available
as a tarball or accessible
from through the GitHub project MC_cython.
A file make.sh performs all the compilations, while compare.py
runs all methods and prints out the CPU time required by each method,
normalized by the fastest approach. The results for \( N=450,000 \)
are listed below (MacBook Air
running Ubuntu in a VMWare Fusion virtual machine).
| Method | Timing |
|---|---|
| C program | 1.0 |
| Cython stdlib.rand | 1.2 |
| Cython numpy.random | 1.2 |
| C via f2py | 1.2 |
| C via Cython | 1.2 |
| vectorized Python, version 1 | 1.9 |
| Cython random.randint | 33.6 |
| plain Python | 37.7 |
| vectorized Python, version 2 | 105.0 |
The CPU time of the plain Python version was 10 s, which is reasonably
fast for obtaining a fairly accurate result in this problem. The
lesson learned is therefore that a Monte Carlo simulation can be
implemented in plain Python first. If more speed is needed, one can
just add type information and create a Cython code. Studying the HTML
file with what Cython manages to translate to C may give hints about
how successful the Cython code is and point to optimizations, like
avoiding the call to random.randint in the present case. Optimal
Cython code runs here at approximately the same speed as calling a
handwritten C function with the time-consuming loops. It is
to be noticed that the stand-alone C program here ran faster than
calling C from Python, probably because the amount of calculations is
not large enough to make the overhead of calling C negligible.
Vectorized Python do give a great speed-up compared to plain loops in Python, if done correctly, but the efficiency is not on par with Cython or handwritten C. Even more important is the fact that vectorized code is not at all as readable as the algorithm expressed in plain Python, Cython, or C. Cython therefore provides a very attractive combination of readability, ease of programming, and high speed.
This chapter is taken from the book A Primer on Scientific Programming with Python by H. P. Langtangen, 4th edition, Springer, 2014.