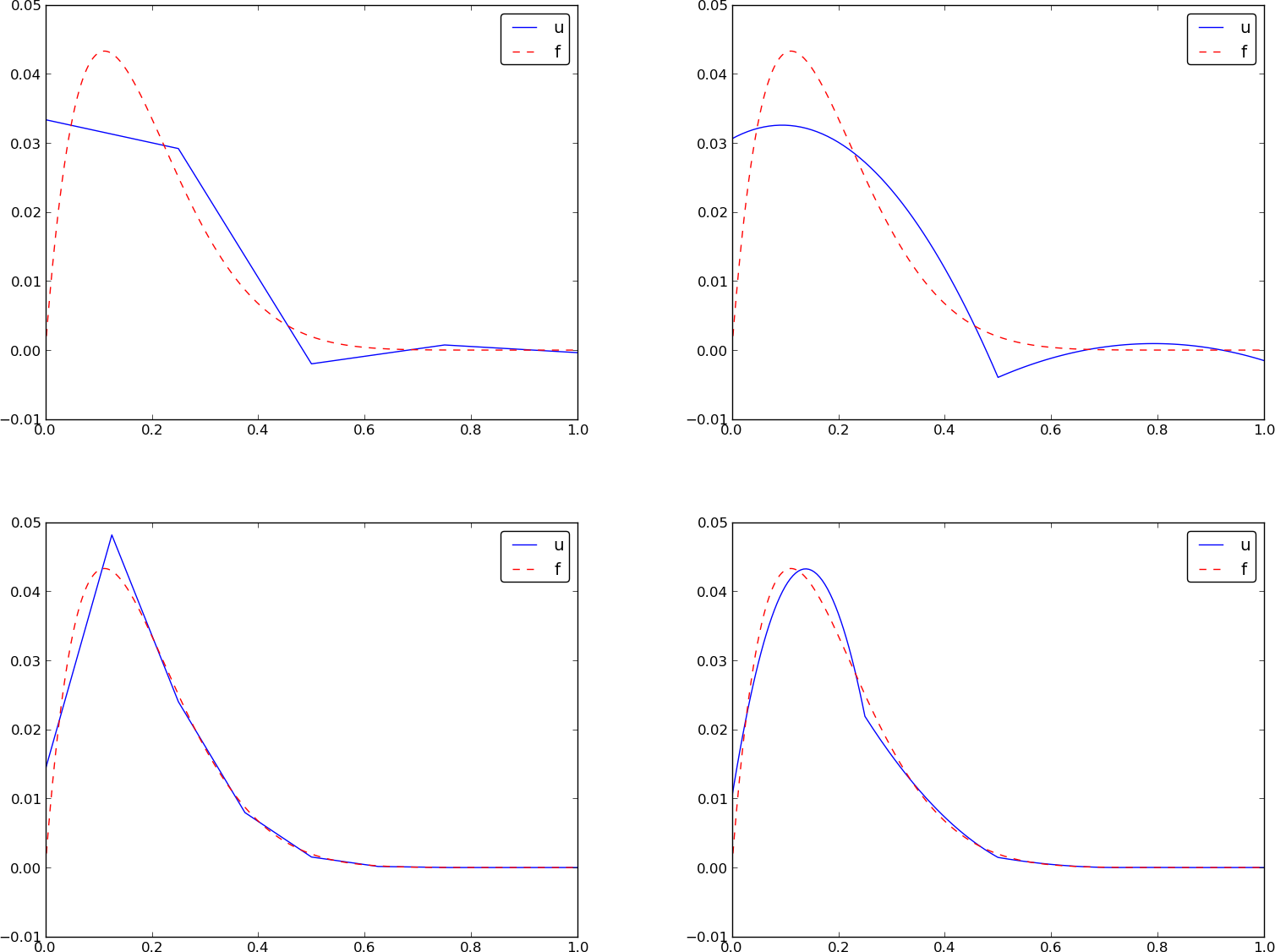
Figure 31: Comparison of the finite element approximations: 4 P1 elements with 5 nodes (upper left), 2 P2 elements with 5 nodes (upper right), 8 P1 elements with 9 nodes (lower left), and 4 P2 elements with 9 nodes (lower right).
Based on the experience from the previous example, it makes sense to write some code to automate the analytical integration process for any choice of finite element basis functions. In addition, we can automate the assembly process and the solution of the linear system. Another advantage is that the code for these purposes document all details of all steps in the finite element computational machinery. The complete code can be found in the module file fe_approx1D.py.
First we need a Python function for
defining \( \refphi_r(X) \) in terms of a Lagrange polynomial
of degree d:
import sympy as sym
import numpy as np
def basis(d, point_distribution='uniform', symbolic=False):
"""
Return all local basis function phi as functions of the
local point X in a 1D element with d+1 nodes.
If symbolic=True, return symbolic expressions, else
return Python functions of X.
point_distribution can be 'uniform' or 'Chebyshev'.
"""
X = sym.symbols('X')
if d == 0:
phi_sym = [1]
else:
if point_distribution == 'uniform':
if symbolic:
# Compute symbolic nodes
h = sym.Rational(1, d) # node spacing
nodes = [2*i*h - 1 for i in range(d+1)]
else:
nodes = np.linspace(-1, 1, d+1)
elif point_distribution == 'Chebyshev':
# Just numeric nodes
nodes = Chebyshev_nodes(-1, 1, d)
phi_sym = [Lagrange_polynomial(X, r, nodes)
for r in range(d+1)]
# Transform to Python functions
phi_num = [sym.lambdify([X], phi_sym[r], modules='numpy')
for r in range(d+1)]
return phi_sym if symbolic else phi_num
def Lagrange_polynomial(x, i, points):
p = 1
for k in range(len(points)):
if k != i:
p *= (x - points[k])/(points[i] - points[k])
return p
Observe how we construct the phi_sym list to be
symbolic expressions for \( \refphi_r(X) \) with X as a
Symbol object from sympy. Also note that the
Lagrange_polynomial function (here simply copied
from the section Fourier series)
works with both symbolic and numeric variables.
Now we can write the function that computes the element matrix
with a list of symbolic expressions for \( \basphi_r \)
(phi = basis(d, symbolic=True)):
def element_matrix(phi, Omega_e, symbolic=True):
n = len(phi)
A_e = sym.zeros((n, n))
X = sym.Symbol('X')
if symbolic:
h = sym.Symbol('h')
else:
h = Omega_e[1] - Omega_e[0]
detJ = h/2 # dx/dX
for r in range(n):
for s in range(r, n):
A_e[r,s] = sym.integrate(phi[r]*phi[s]*detJ, (X, -1, 1))
A_e[s,r] = A_e[r,s]
return A_e
In the symbolic case (symbolic is True),
we introduce the element length as a symbol
h in the computations. Otherwise, the real numerical value
of the element interval Omega_e
is used and the final matrix elements are numbers,
not symbols.
This functionality can be demonstrated:
>>> from fe_approx1D import *
>>> phi = basis(d=1, symbolic=True)
>>> phi
[-X/2 + 1/2, X/2 + 1/2]
>>> element_matrix(phi, Omega_e=[0.1, 0.2], symbolic=True)
[h/3, h/6]
[h/6, h/3]
>>> element_matrix(phi, Omega_e=[0.1, 0.2], symbolic=False)
[0.0333333333333333, 0.0166666666666667]
[0.0166666666666667, 0.0333333333333333]
The computation of the element vector is done by a similar procedure:
def element_vector(f, phi, Omega_e, symbolic=True):
n = len(phi)
b_e = sym.zeros((n, 1))
# Make f a function of X
X = sym.Symbol('X')
if symbolic:
h = sym.Symbol('h')
else:
h = Omega_e[1] - Omega_e[0]
x = (Omega_e[0] + Omega_e[1])/2 + h/2*X # mapping
f = f.subs('x', x) # substitute mapping formula for x
detJ = h/2 # dx/dX
for r in range(n):
b_e[r] = sym.integrate(f*phi[r]*detJ, (X, -1, 1))
return b_e
Here we need to replace the symbol x in the expression for f
by the mapping formula such that f can be integrated
in terms of \( X \), cf. the formula
\( \tilde b^{(e)}_{r} = \int_{-1}^1 f(x(X))\refphi_r(X)\frac{h}{2}\dX \).
The integration in the element matrix function involves only products
of polynomials, which sympy can easily deal with, but for the
right-hand side sympy may face difficulties with certain types of
expressions f. The result of the integral is then an Integral
object and not a number or expression
as when symbolic integration is successful.
It may therefore be wise to introduce a fall back on numerical
integration. The symbolic integration can also spend considerable time
before reaching an unsuccessful conclusion, so we may also introduce a parameter
symbolic to turn symbolic integration on and off:
def element_vector(f, phi, Omega_e, symbolic=True):
...
if symbolic:
I = sym.integrate(f*phi[r]*detJ, (X, -1, 1))
if not symbolic or isinstance(I, sym.Integral):
h = Omega_e[1] - Omega_e[0] # Ensure h is numerical
detJ = h/2
integrand = sym.lambdify([X], f*phi[r]*detJ)
I = sym.mpmath.quad(integrand, [-1, 1])
b_e[r] = I
...
Numerical integration requires that the symbolic
integrand is converted
to a plain Python function (integrand) and that
the element length h is a real number.
The complete algorithm for computing and assembling the elementwise contributions takes the following form
def assemble(nodes, elements, phi, f, symbolic=True):
N_n, N_e = len(nodes), len(elements)
if symbolic:
A = sym.zeros((N_n, N_n))
b = sym.zeros((N_n, 1)) # note: (N_n, 1) matrix
else:
A = np.zeros((N_n, N_n))
b = np.zeros(N_n)
for e in range(N_e):
Omega_e = [nodes[elements[e][0]], nodes[elements[e][-1]]]
A_e = element_matrix(phi, Omega_e, symbolic)
b_e = element_vector(f, phi, Omega_e, symbolic)
for r in range(len(elements[e])):
for s in range(len(elements[e])):
A[elements[e][r],elements[e][s]] += A_e[r,s]
b[elements[e][r]] += b_e[r]
return A, b
The nodes and elements variables represent the finite
element mesh as explained earlier.
Given the coefficient matrix A and the right-hand side b,
we can compute the coefficients \( \sequencej{c} \) in the expansion
\( u(x)=\sum_jc_j\basphi_j \) as the solution vector c of the linear
system:
if symbolic:
c = A.LUsolve(b)
else:
c = np.linalg.solve(A, b)
When A and b are sympy arrays,
the solution procedure implied by A.LUsolve is symbolic.
Otherwise, A and b are numpy arrays and a standard
numerical solver is called.
The symbolic version is suited for small problems only
(small \( N \) values) since the calculation time becomes prohibitively large
otherwise. Normally, the symbolic integration will be more time
consuming in small problems than the symbolic solution of the linear system.
We can exemplify the use of assemble on the computational
case from the section Calculating the linear system with
two P1 elements (linear basis functions) on the domain \( \Omega=[0,1] \).
Let us first work with a symbolic element length:
>>> h, x = sym.symbols('h x')
>>> nodes = [0, h, 2*h]
>>> elements = [[0, 1], [1, 2]]
>>> phi = basis(d=1, symbolic=True)
>>> f = x*(1-x)
>>> A, b = assemble(nodes, elements, phi, f, symbolic=True)
>>> A
[h/3, h/6, 0]
[h/6, 2*h/3, h/6]
[ 0, h/6, h/3]
>>> b
[ h**2/6 - h**3/12]
[ h**2 - 7*h**3/6]
[5*h**2/6 - 17*h**3/12]
>>> c = A.LUsolve(b)
>>> c
[ h**2/6]
[12*(7*h**2/12 - 35*h**3/72)/(7*h)]
[ 7*(4*h**2/7 - 23*h**3/21)/(2*h)]
As an alternative to the least squares formulation,
we may compute the c vector based on
the interpolation method from the section The interpolation (or collocation) method,
using finite element basis functions.
Choosing the nodes as interpolation points, the method can be written as
$$ u(\xno{i}) = \sum_{j\in\If} c_j\basphi_j(\xno{i}) = f(\xno{i}),\quad
i\in\If\tp$$
The coefficient matrix \( A_{i,j}=\basphi_j(\xno{i}) \) becomes
the identity matrix because basis function number \( j \) vanishes
at all nodes, except node \( i \): \( \basphi_j(\xno{i})=\delta_{ij} \).
Therefore, \( c_i = f(\xno{i}) \).
The associated sympy calculations are
>>> fn = sym.lambdify([x], f)
>>> c = [fn(xc) for xc in nodes]
>>> c
[0, h*(1 - h), 2*h*(1 - 2*h)]
These expressions are much simpler than those based on least squares or projection in combination with finite element basis functions. However, which of the two methods that is most appropriate for a given task is problem-dependent, so we need both methods in our toolbox.
The numerical computations corresponding to the
symbolic ones in the section Example on computing symbolic approximations
(still done by sympy and the assemble function) go as follows:
>>> nodes = [0, 0.5, 1]
>>> elements = [[0, 1], [1, 2]]
>>> phi = basis(d=1, symbolic=True)
>>> x = sym.Symbol('x')
>>> f = x*(1-x)
>>> A, b = assemble(nodes, elements, phi, f, symbolic=False)
>>> A
[ 0.166666666666667, 0.0833333333333333, 0]
[0.0833333333333333, 0.333333333333333, 0.0833333333333333]
[ 0, 0.0833333333333333, 0.166666666666667]
>>> b
[ 0.03125]
[0.104166666666667]
[ 0.03125]
>>> c = A.LUsolve(b)
>>> c
[0.0416666666666666]
[ 0.291666666666667]
[0.0416666666666666]
The fe_approx1D module contains functions for generating the
nodes and elements lists for equal-sized elements with
any number of nodes per element. The coordinates in nodes
can be expressed either through the element length symbol h
(symbolic=True) or by real numbers (symbolic=False):
nodes, elements = mesh_uniform(N_e=10, d=3, Omega=[0,1],
symbolic=True)
There is also a function
def approximate(f, symbolic=False, d=1, N_e=4, filename='tmp.pdf'):
which computes a mesh with N_e elements, basis functions of
degree d, and approximates a given symbolic expression
f by a finite element expansion \( u(x) = \sum_jc_j\basphi_j(x) \).
When symbolic is False, \( u(x) = \sum_jc_j\basphi_j(x) \)
can be computed at a (large)
number of points and plotted together with \( f(x) \). The construction
of \( u \) points from the solution vector c is done
elementwise by evaluating \( \sum_rc_r\refphi_r(X) \) at a (large)
number of points in each element in the local coordinate system,
and the discrete \( (x,u) \) values on
each element are stored in separate arrays that are finally
concatenated to form a global array for \( x \) and for \( u \).
The details are found in the u_glob function in
fe_approx1D.py.
Let us first see how the global matrix looks like if we assemble
symbolic element matrices, expressed in terms of h, from
several elements:
>>> d=1; N_e=8; Omega=[0,1] # 8 linear elements on [0,1]
>>> phi = basis(d)
>>> f = x*(1-x)
>>> nodes, elements = mesh_symbolic(N_e, d, Omega)
>>> A, b = assemble(nodes, elements, phi, f, symbolic=True)
>>> A
[h/3, h/6, 0, 0, 0, 0, 0, 0, 0]
[h/6, 2*h/3, h/6, 0, 0, 0, 0, 0, 0]
[ 0, h/6, 2*h/3, h/6, 0, 0, 0, 0, 0]
[ 0, 0, h/6, 2*h/3, h/6, 0, 0, 0, 0]
[ 0, 0, 0, h/6, 2*h/3, h/6, 0, 0, 0]
[ 0, 0, 0, 0, h/6, 2*h/3, h/6, 0, 0]
[ 0, 0, 0, 0, 0, h/6, 2*h/3, h/6, 0]
[ 0, 0, 0, 0, 0, 0, h/6, 2*h/3, h/6]
[ 0, 0, 0, 0, 0, 0, 0, h/6, h/3]
The reader is encouraged to assemble the element matrices by hand and verify this result, as this exercise will give a hands-on understanding of what the assembly is about. In general we have a coefficient matrix that is tridiagonal: $$ \begin{equation} A = \frac{h}{6} \left( \begin{array}{cccccccccc} 2 & 1 & 0 &\cdots & \cdots & \cdots & \cdots & \cdots & 0 \\ 1 & 4 & 1 & \ddots & & & & & \vdots \\ 0 & 1 & 4 & 1 & \ddots & & & & \vdots \\ \vdots & \ddots & & \ddots & \ddots & 0 & & & \vdots \\ \vdots & & \ddots & \ddots & \ddots & \ddots & \ddots & & \vdots \\ \vdots & & & 0 & 1 & 4 & 1 & \ddots & \vdots \\ \vdots & & & & \ddots & \ddots & \ddots &\ddots & 0 \\ \vdots & & & & &\ddots & 1 & 4 & 1 \\ 0 &\cdots & \cdots &\cdots & \cdots & \cdots & 0 & 1 & 2 \end{array} \right) \tag{88} \end{equation} $$
The structure of the right-hand side is more difficult to reveal since it involves an assembly of elementwise integrals of \( f(x(X))\refphi_r(X)h/2 \), which obviously depend on the particular choice of \( f(x) \). Numerical integration can give some insight into the nature of the right-hand side. For this purpose it is easier to look at the integration in \( x \) coordinates, which gives the general formula (65). For equal-sized elements of length \( h \), we can apply the Trapezoidal rule at the global node points to arrive at $$ b_i = h\left( \half \basphi_i(\xno{0})f(\xno{0}) + \half \basphi_i(\xno{N})f(\xno{N}) + \sum_{j=1}^{N-1} \basphi_i(\xno{j})f(\xno{j})\right), $$ which leads to $$ \begin{equation} b_i = \left\lbrace\begin{array}{ll} \half hf(x_i),& i=0\hbox{ or }i=N,\\ h f(x_i), & 1 \leq i \leq N-1 \end{array}\right. \tag{89} \end{equation} $$ The reason for this simple formula is just that \( \basphi_i \) is either 0 or 1 at the nodes and 0 at all but one of them.
Going to P2 elements (d=2) leads
to the element matrix
$$
\begin{equation}
A^{(e)} = \frac{h}{30}
\left(\begin{array}{ccc}
4 & 2 & -1\\
2 & 16 & 2\\
-1 & 2 & 4
\end{array}\right)
\tag{90}
\end{equation}
$$
and the following global matrix, assembled here from four elements:
$$
\begin{equation}
A = \frac{h}{30}
\left(
\begin{array}{ccccccccc}
4 & 2 & - 1 & 0
& 0 & 0 & 0 & 0 & 0\\
2 & 16 & 2
& 0 & 0 & 0 & 0 & 0 & 0\\- 1 & 2 &
8 & 2 & - 1 & 0 & 0 & 0 &
0\\0 & 0 & 2 & 16 & 2 & 0 & 0
& 0 & 0\\0 & 0 & - 1 & 2 & 8
& 2 & - 1 & 0 & 0\\0 & 0 & 0 & 0 &
2 & 16 & 2 & 0 & 0\\0 & 0 & 0
& 0 & - 1 & 2 & 8 &
2 & - 1\\0 & 0 & 0 & 0 & 0 & 0 &
2 & 16 & 2\\0 & 0 & 0 & 0 & 0
& 0 & - 1 & 2 & 4
\end{array}
\right)
\tag{91}
\end{equation}
$$
In general, for \( i \) odd we have the nonzeroes
$$
\begin{equation*} A_{i,i-2} = -1,\quad A_{i-1,i}=2,\quad A_{i,i} = 8,\quad A_{i+1,i}=2,
\quad A_{i+2,i}=-1,\end{equation*}
$$
multiplied by \( h/30 \), and for \( i \) even we have the nonzeros
$$
\begin{equation*} A_{i-1,i}=2,\quad A_{i,i} = 16,\quad A_{i+1,i}=2,\end{equation*}
$$
multiplied by \( h/30 \). The rows with odd numbers correspond to
nodes at the element boundaries and get contributions from two
neighboring elements in the assembly process,
while the even numbered rows correspond to
internal nodes in the elements where only one element contributes
to the values in the global matrix.
With the aid of the approximate function in the fe_approx1D
module we can easily investigate the quality of various finite element
approximations to some given functions. Figure 31
shows how linear and quadratic elements approximate the polynomial
\( f(x)=x(1-x)^8 \) on \( \Omega =[0,1] \), using equal-sized elements.
The results arise from the program
import sympy as sym
from fe_approx1D import approximate
x = sym.Symbol('x')
approximate(f=x*(1-x)**8, symbolic=False, d=1, N_e=4)
approximate(f=x*(1-x)**8, symbolic=False, d=2, N_e=2)
approximate(f=x*(1-x)**8, symbolic=False, d=1, N_e=8)
approximate(f=x*(1-x)**8, symbolic=False, d=2, N_e=4)
The quadratic functions are seen to be better than the linear ones for the same value of \( N \), as we increase \( N \). This observation has some generality: higher degree is not necessarily better on a coarse mesh, but it is as we refine the mesh.
Figure 31: Comparison of the finite element approximations: 4 P1 elements with 5 nodes (upper left), 2 P2 elements with 5 nodes (upper right), 8 P1 elements with 9 nodes (lower left), and 4 P2 elements with 9 nodes (lower right).

Some of the examples in the preceding section took several minutes to
compute, even on small meshes consisting of up to eight elements.
The main explanation for slow computations is unsuccessful
symbolic integration: sympy may use a lot of energy on
integrals like \( \int f(x(X))\refphi_r(X)h/2 \dx \) before
giving up, and the program then resorts to numerical integration.
Codes that can deal with a large number of basis functions and
accept flexible choices of \( f(x) \) should compute all integrals
numerically and replace the matrix objects from sympy by
the far more efficient array objects from numpy.
There is also another (potential) reason for slow code: the solution algorithm for the linear system performs much more work than necessary. Most of the matrix entries \( A_{i,j} \) are zero, because \( (\basphi_i,\basphi_j)=0 \) unless \( i \) and \( j \) are nodes in the same element. In 1D problems, we do not need to store or compute with these zeros when solving the linear system, but that requires solution methods adapted to the kind of matrices produced by the finite element approximations.
A matrix whose majority of entries are zeros, is known as a sparse matrix. Utilizing sparsity in software dramatically decreases the storage demands and the CPU-time needed to compute the solution of the linear system. This optimization is not very critical in 1D problems where modern computers can afford computing with all the zeros in the complete square matrix, but in 2D and especially in 3D, sparse matrices are fundamental for feasible finite element computations. One of the advantageous features of the finite element method is that it produces very sparse matrices. The reason is that the basis functions have local support such that the product of two basis functions, as typically met in integrals, is mostly zero.
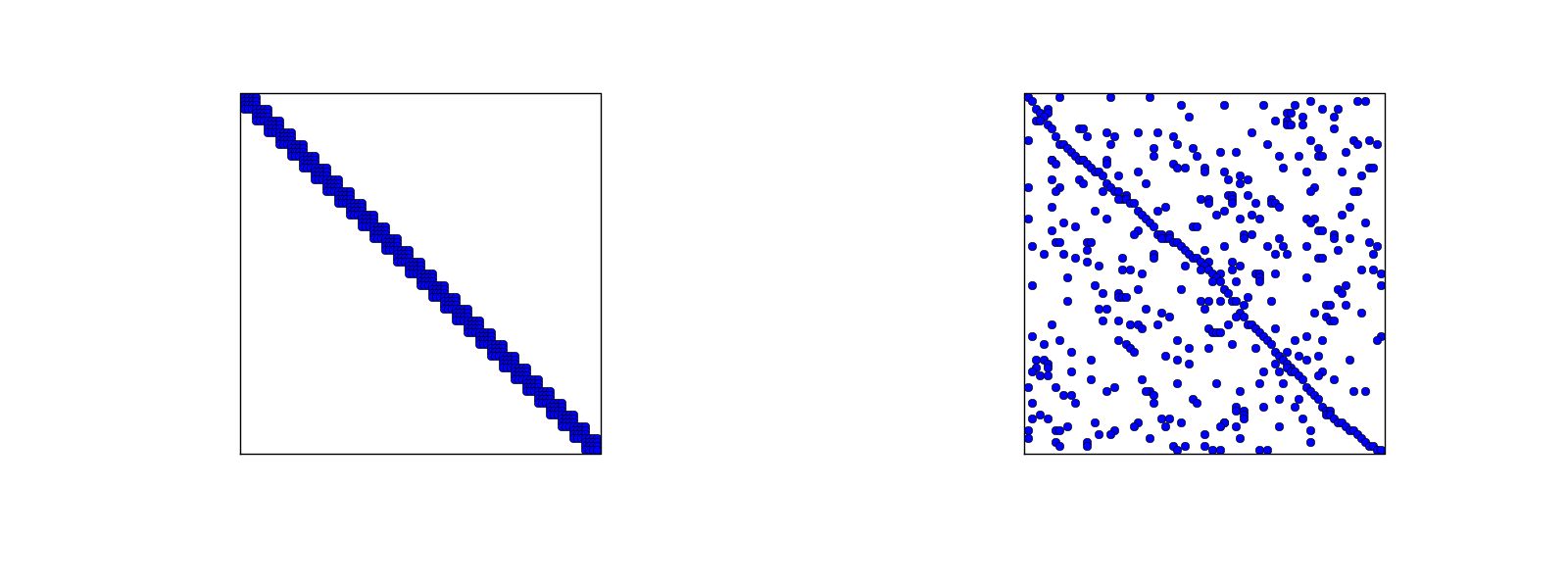
Using a numbering of nodes and elements from left to right over a 1D domain, the assembled coefficient matrix has only a few diagonals different from zero. More precisely, \( 2d+1 \) diagonals around the main diagonal are different from zero. With a different numbering of global nodes, say a random ordering, the diagonal structure is lost, but the number of nonzero elements is unaltered. Figures 32 and 33 exemplify sparsity patterns.
Figure 32: Matrix sparsity pattern for left-to-right numbering (left) and random numbering (right) of nodes in P1 elements.
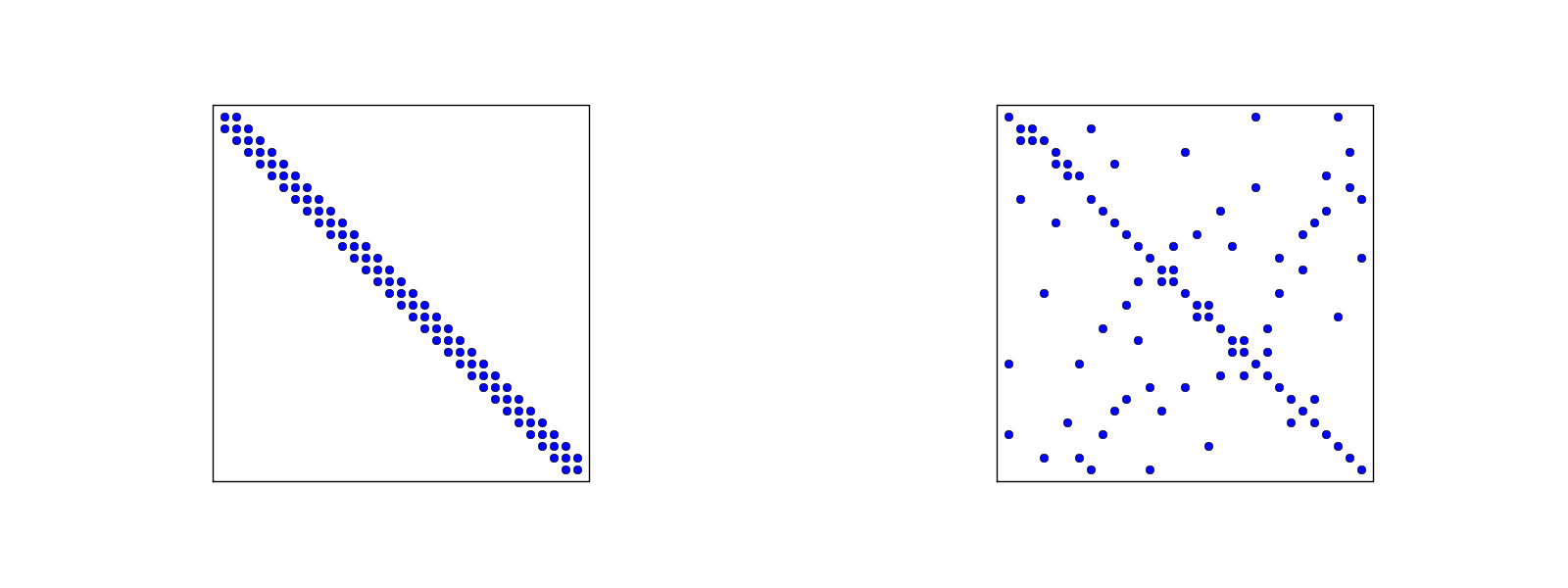
Figure 33: Matrix sparsity pattern for left-to-right numbering (left) and random numbering (right) of nodes in P3 elements.
The scipy.sparse library supports creation of sparse matrices
and linear system solution.
scipy.sparse.diags for matrix defined via diagonalsscipy.sparse.dok_matrix for matrix incrementally defined via index pairs \( (i,j) \)dok_matrix object is most convenient for finite element computations.
This sparse matrix format is called DOK, which stands for Dictionary Of Keys:
the implementation is basically a dictionary (hash) with the
entry indices (i,j) as keys.
Rather than declaring A = np.zeros((N_n, N_n)), a DOK sparse
matrix is created by
import scipy.sparse
A = scipy.sparse.dok_matrix((N_n, N_n))
When there is any need to add or set some matrix entry i,j, just do
A[i,j] = entry
# or
A[i,j] += entry
The indexing creates the matrix entry on the fly, and only the nonzero entries in the matrix will be stored.
To solve a system with right-hand side b (one-dimensional numpy
array) with a sparse coefficient matrix A, we must use some kind of
a sparse linear system solver. The safest choice is a method based on
sparse Gaussian elimination:
import scipy.sparse.linalg
c = scipy.sparse.linalg.spsolve(A.tocsr(), b, use_umfpack=True)
The call A.tocsr() is not strictly needed (a warning is issued
otherwise), but ensures that the solution algorithm can efficiently
work with a copy of the sparse matrix in Compressed Sparse Row (CSR) format.
An advantage of the scipy.sparse.diags matrix over the DOK format is
that the former allows vectorized assignment to the matrix.
Vectorization is possible for approximation problems when all elements
are of the same type. However, when solving differential equations,
vectorization is much more difficult. It also appears that the DOK
sparse matrix format available in the scipy.sparse package is fast
enough even for big 1D problems on today's laptops, so the need for
improving efficiency occurs in 2D and 3D problems, but then the
complexity of the mesh favors the DOK format.