Creating user interfaces
Accessing command-line arguments
Reading a sequence of command-line arguments
Implementation
Working with an argument parser
Reading option-values pairs
A graphical user interface
The Parampool package
Making a compute function
The hard part of the compute function: the HTML code
How to embed a PNG plot in HTML code
Generating the user interface
Running the web application
More advanced use
Computing convergence rates
Estimating the convergence rate \( r \)
Implementation
Execution
Debugging via convergence rates
Software engineering
Making a module
Sketch of the module
Test block
Extended test block
Prefixing imported functions by the module name
Downside of module prefix notation
Doctests
Running doctests
Unit testing with nose
Basic use of nose
Example on a nose test in the source code
Example on a nose test in a separate file
The habit of writing nose tests
Purpose of a test function: raise AssertionError if failure
Advantages of nose
Demonstrating nose (ideas)
Demonstrating nose (code)
Floats as test results require careful comparison
Test of wrong use
Test of convergence rates
Classical unit testing with unittest
Basic use of unittest
Demonstration of unittest
Implementing simple problem and solver classes
What to learn
The problem class
Improved problem class
The solver class
The visualizer class
Combing the classes
Implementing more advanced problem and solver classes
A generic class for parameters
The problem class
The solver class
The visualizer class
Performing scientific experiments
Model problem and numerical solution method
Plan for the experiments
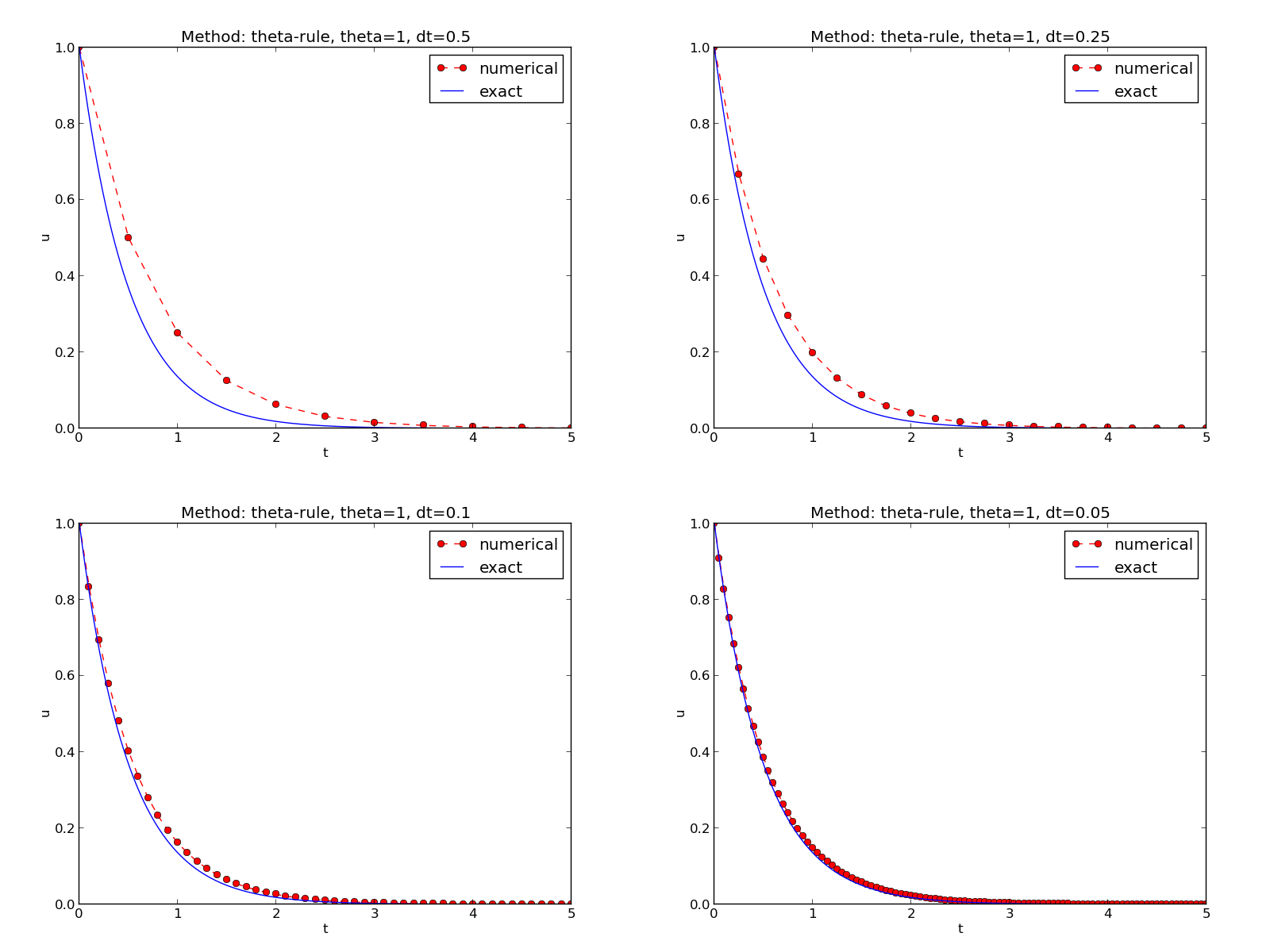
Typical plot summarizing the results
Script code
Comments to the code
Interpreting output from other programs
Code for grabbing output from another program
Code for interpreting the grabbed output
Making a report
Publishing a complete project
sys.argvsys.argv[0] is the programsys.argv[1:] holds the command-line arguments--option value pairs on the command line (with default values)
Terminal> python myprog.py 1.5 2 0.5 0.8 0.4
Terminal> python myprog.py --I 1.5 --a 2 --dt 0.8 0.4
The program decay_plot.py needs this input:
makeplot)
Terminal> python decay_cml.py 1.5 2 0.5 0.8 0.4
import sys
def read_command_line():
if len(sys.argv) < 6:
print 'Usage: %s I a T on/off dt1 dt2 dt3 ...' % \
sys.argv[0]; sys.exit(1) # abort
I = float(sys.argv[1])
a = float(sys.argv[2])
T = float(sys.argv[3])
makeplot = sys.argv[4] in ('on', 'True')
dt_values = [float(arg) for arg in sys.argv[5:]]
return I, a, T, makeplot, dt_values
Note:
sys.argv[i] is always a stringfloat for computations[expression for e in somelist]
Set option-value pairs on the command line if the default value is not suitable:
Terminal> python decay_argparse.py --I 1.5 --a 2 --dt 0.8 0.4
Code:
def define_command_line_options():
import argparse
parser = argparse.ArgumentParser()
parser.add_argument('--I', '--initial_condition', type=float,
default=1.0, help='initial condition, u(0)',
metavar='I')
parser.add_argument('--a', type=float,
default=1.0, help='coefficient in ODE',
metavar='a')
parser.add_argument('--T', '--stop_time', type=float,
default=1.0, help='end time of simulation',
metavar='T')
parser.add_argument('--makeplot', action='store_true',
help='display plot or not')
parser.add_argument('--dt', '--time_step_values', type=float,
default=[1.0], help='time step values',
metavar='dt', nargs='+', dest='dt_values')
return parser
(metavar is the symbol used in help output)
argparse.ArgumentParser parses the command-line arguments:
def read_command_line():
parser = define_command_line_options()
args = parser.parse_args()
print 'I={}, a={}, T={}, makeplot={}, dt_values={}'.format(
args.I, args.a, args.T, args.makeplot, args.dt_values)
return args.I, args.a, args.T, args.makeplot, args.dt_values
Complete program: decay_argparse.py.

Normally very much programming required - and much competence on graphical user interfaces.
Here: use a tool to automatically create it in a few minutes (!)
The forthcoming material aims at those with particular interest in equipping their programs with a GUI - others can safely skip it.
main function carries out simulations and plotting for a
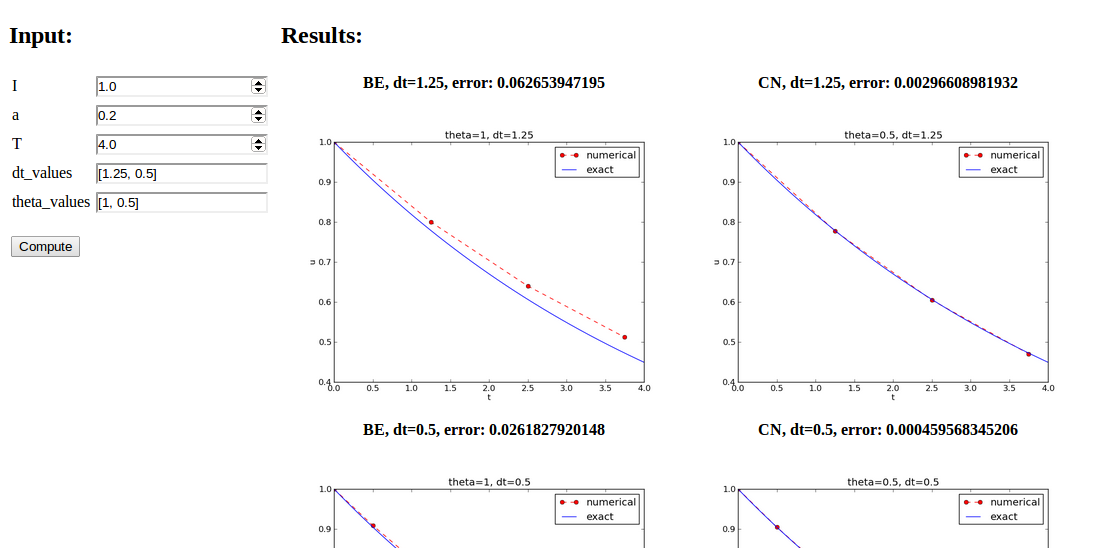
series of \( \Delta t \) valuesparampool functionalitymain_GUI:
def main_GUI(I=1.0, a=.2, T=4.0,
dt_values=[1.25, 0.75, 0.5, 0.1],
theta_values=[0, 0.5, 1]):
explore solves the problem, makes a plot, computes the
error and returns appropriate HTML code with the plot. Embed
error and plots in a table:
def main_GUI(I=1.0, a=.2, T=4.0,
dt_values=[1.25, 0.75, 0.5, 0.1],
theta_values=[0, 0.5, 1]):
# Build HTML code for web page. Arrange plots in columns
# corresponding to the theta values, with dt down the rows
theta2name = {0: 'FE', 1: 'BE', 0.5: 'CN'}
html_text = '<table>\n'
for dt in dt_values:
html_text += '<tr>\n'
for theta in theta_values:
E, html = explore(I, a, T, dt, theta, makeplot=True)
html_text += """
<td>
<center><b>%s, dt=%g, error: %s</b></center><br>
%s
</td>
""" % (theta2name[theta], dt, E, html)
html_text += '</tr>\n'
html_text += '</table>\n'
return html_text
In explore:
import matplotlib.pyplot as plt
...
# plot
plt.plot(t, u, r-')
plt.xlabel('t')
plt.ylabel('u')
...
from parampool.utils import save_png_to_str
html_text = save_png_to_str(plt, plotwidth=400)
If you know HTML, you can return more sophisticated layout etc.
Make a file decay_GUI_generate.py:
from parampool.generator.flask import generate
from decay_GUI import main
generate(main,
output_controller='decay_GUI_controller.py',
output_template='decay_GUI_view.py',
output_model='decay_GUI_model.py')
Running decay_GUI_generate.py results in
decay_GUI_model.py defines HTML widgets to be used to set
input data in the web interface,templates/decay_GUI_views.py defines the layout of the web page,decay_GUI_controller.py runs the web application.decay_GUI_controller.py
and there is no need to look into any of these files!
Start the GUI
Terminal> python decay_GUI_controller.py
Open a web browser at 127.0.0.1:5000

Frequent assumption on the relation between the numerical error \( E \) and some discretization parameter \( \Delta t \): $$ \begin{equation} E = C\Delta t^r, \label{decay:E:dt} \end{equation} $$
Perform numerical experiments: \( (\Delta t_i, E_i) \), \( i=0,\ldots,m-1 \). Two methods for finding \( r \) (and \( C \)):
Method 2 is best.
Compute \( r_0, r_1, \ldots, r_{m-2} \):
from math import log
def main():
I, a, T, makeplot, dt_values = read_command_line()
r = {} # estimated convergence rates
for theta in 0, 0.5, 1:
E_values = []
for dt in dt_values:
E = explore(I, a, T, dt, theta, makeplot=False)
E_values.append(E)
# Compute convergence rates
m = len(dt_values)
r[theta] = [log(E_values[i-1]/E_values[i])/
log(dt_values[i-1]/dt_values[i])
for i in range(1, m, 1)]
for theta in r:
print '\nPairwise convergence rates for theta=%g:' % theta
print ' '.join(['%.2f' % r_ for r_ in r[theta]])
return r
Complete program: decay_convrate.py.
Terminal> python decay_convrate.py --dt 0.5 0.25 0.1 0.05 0.025 0.01
...
Pairwise convergence rates for theta=0:
1.33 1.15 1.07 1.03 1.02
Pairwise convergence rates for theta=0.5:
2.14 2.07 2.03 2.01 2.01
Pairwise convergence rates for theta=1:
0.98 0.99 0.99 1.00 1.00
Verify that \( r \) has the expected value!
Potential bug: missing a in the denominator,
u[n+1] = (1 - (1-theta)*a*dt)/(1 + theta*dt)*u[n]
Running decay_convrate.py gives same rates.
Why? The value of \( a \)... (\( a=1 \))
0 and 1 are bad values in tests!
Better:
Terminal> python decay_convrate.py --a 2.1 --I 0.1 \
--dt 0.5 0.25 0.1 0.05 0.025 0.01
...
Pairwise convergence rates for theta=0:
1.49 1.18 1.07 1.04 1.02
Pairwise convergence rates for theta=0.5:
-1.42 -0.22 -0.07 -0.03 -0.01
Pairwise convergence rates for theta=1:
0.21 0.12 0.06 0.03 0.01
Forward Euler works...because \( \theta=0 \) hides the bug.
This bug gives \( r\approx 0 \):
u[n+1] = ((1-theta)*a*dt)/(1 + theta*dt*a)*u[n]
Goal: make more professional numerical software.
Topics:
solver)import)solververify_three_stepsverify_discrete_solutionexploredefine_command_line_optionsread_command_linemain (with convergence rates)verify_convergence_ratedecay_mod, filename: decay_mod.py.
from numpy import *
from matplotlib.pyplot import *
import sys
def solver(I, a, T, dt, theta):
...
def verify_three_steps():
...
def verify_exact_discrete_solution():
...
def u_exact(t, I, a):
...
def explore(I, a, T, dt, theta=0.5, makeplot=True):
...
def define_command_line_options():
...
def read_command_line(use_argparse=True):
...
def main():
...
That is! It's a module decay_mod in file decay_mod.py.
Usage in some other program:
from decay_mod import solver
u, t = solver(I=1.0, a=3.0, T=3, dt=0.01, theta=0.5)
At the end of a module it is common to include a test block:
if __name__ == '__main__':
main()
Note:
decay_mod is imported, __name__ is decay_mod.decay_mod.py is run, __name__ is __main__.
if __name__ == '__main__':
if 'verify' in sys.argv:
if verify_three_steps() and verify_discrete_solution():
pass # ok
else:
print 'Bug in the implementation!'
elif 'verify_rates' in sys.argv:
sys.argv.remove('verify_rates')
if not '--dt' in sys.argv:
print 'Must assign several dt values'
sys.exit(1) # abort
if verify_convergence_rate():
pass
else:
print 'Bug in the implementation!'
else:
# Perform simulations
main()
from numpy import *
from matplotlib.pyplot import *
This imports a large number of names (sin, exp, linspace, plot, ...).
Confusion: is a function from numpy? Or matplotlib.pyplot?
Alternative (recommended) import:
import numpy
import matplotlib.pyplot
Now we need to prefix functions with module name:
t = numpy.linspace(0, T, Nt+1)
u_e = I*numpy.exp(-a*t)
matplotlib.pyplot.plot(t, u_e)
Common standard:
import numpy as np
import matplotlib.pyplot as plt
t = np.linspace(0, T, Nt+1)
u_e = I*np.exp(-a*t)
plt.plot(t, u_e)
A math line like \( e^{-at}\sin(2\pi t) \) gets cluttered with module names,
numpy.exp(-a*t)*numpy.sin(2(numpy.pi*t)
# or
np.exp(-a*t)*np.sin(2*np.pi*t)
Solution (much used in this course): do two imports
import numpy as np
from numpy import exp, sin, pi
...
t = np.linspace(0, T, Nt+1)
u_e = exp(-a*t)*sin(2*pi*t)
Doc strings can be equipped with interactive Python sessions for demonstrating usage and automatic testing of functions.
def solver(I, a, T, dt, theta):
"""
Solve u'=-a*u, u(0)=I, for t in (0,T] with steps of dt.
>>> u, t = solver(I=0.8, a=1.2, T=4, dt=0.5, theta=0.5)
>>> for t_n, u_n in zip(t, u):
... print 't=%.1f, u=%.14f' % (t_n, u_n)
t=0.0, u=0.80000000000000
t=0.5, u=0.43076923076923
t=1.0, u=0.23195266272189
t=1.5, u=0.12489758761948
t=2.0, u=0.06725254717972
t=2.5, u=0.03621291001985
t=3.0, u=0.01949925924146
t=3.5, u=0.01049960113002
t=4.0, u=0.00565363137770
"""
...
Automatic check that the code reproduces the doctest output:
Terminal> python -m doctest decay_mod_doctest.py
Report in case of failure:
Terminal> python -m doctest decay_mod_doctest.py
********************************************************
File "decay_mod_doctest.py", line 12, in decay_mod_doctest....
Failed example:
for t_n, u_n in zip(t, u):
print 't=%.1f, u=%.14f' % (t_n, u_n)
Expected:
t=0.0, u=0.80000000000000
t=0.5, u=0.43076923076923
t=1.0, u=0.23195266272189
t=1.5, u=0.12489758761948
t=2.0, u=0.06725254717972
Got:
t=0.0, u=0.80000000000000
t=0.5, u=0.43076923076923
t=1.0, u=0.23195266272189
t=1.5, u=0.12489758761948
t=2.0, u=0.06725254718756
********************************************************
1 items had failures:
1 of 2 in decay_mod_doctest.solver
***Test Failed*** 1 failures.
Limit the number of digits in the output in doctests! Otherwise, round-off errors on a different machine may ruin the test.
Complete program: decay_mod_doctest.py.
test_.assert functions from the nose.tools module.test*.py.
Very simple module mymod (in file mymod.py):
def double(n):
return 2*n
Write test function in mymod.py:
def double(n):
return 2*n
import nose.tools as nt
def test_double():
result = double(4)
nt.assert_equal(result, 8)
Running
Terminal> nosetests -s mymod
makes the nose tool run all test_*() functions in mymod.py.
Write the test in a separate file, say test_mymod.py:
import nose.tools as nt
import mymod
def test_double():
result = mymod.double(4)
nt.assert_equal(result, 8)
Running
Terminal> nosetests -s
makes the nose tool run all test_*() functions in all files
test*.py in the current directory and in all subdirectories (recursevely)
with names tests or *_tests.
Start with test functions in the source code file. When the file contains many tests, or when you have many source code files, move tests to separate files.
test_*() functions in the moduletest_*() functions, collect them in
tests/test*.py
Alternative ways of raising AssertionError if result is not 8:
import nose.tools as nt
def test_double():
result = ...
nt.assert_equal(result, 8) # alternative 1
assert result == 8 # alternative 2
if result != 8: # alternative 3
raise AssertionError()
nosetests -s)
Aim: test function solver for \( u'=-au \), \( u(0)=I \).
We design three unit tests:
verify* functions.
import nose.tools as nt
import decay_mod_unittest as decay_mod
import numpy as np
def exact_discrete_solution(n, I, a, theta, dt):
"""Return exact discrete solution of the theta scheme."""
dt = float(dt) # avoid integer division
factor = (1 - (1-theta)*a*dt)/(1 + theta*dt*a)
return I*factor**n
def test_exact_discrete_solution():
"""
Compare result from solver against
formula for the discrete solution.
"""
theta = 0.8; a = 2; I = 0.1; dt = 0.8
N = int(8/dt) # no of steps
u, t = decay_mod.solver(I=I, a=a, T=N*dt, dt=dt, theta=theta)
u_de = np.array([exact_discrete_solution(n, I, a, theta, dt)
for n in range(N+1)])
diff = np.abs(u_de - u).max()
nt.assert_almost_equal(diff, 0, delta=1E-14)
nt.assert_almost_equal: compare two floats to some digits
or precision
def test_solver():
"""
Compare result from solver against
precomputed arrays for theta=0, 0.5, 1.
"""
I=0.8; a=1.2; T=4; dt=0.5 # fixed parameters
precomputed = {
't': np.array([ 0. , 0.5, 1. , 1.5, 2. , 2.5,
3. , 3.5, 4. ]),
0.5: np.array(
[ 0.8 , 0.43076923, 0.23195266, 0.12489759,
0.06725255, 0.03621291, 0.01949926, 0.0104996 ,
0.00565363]),
0: ...,
1: ...
}
for theta in 0, 0.5, 1:
u, t = decay_mod.solver(I, a, T, dt, theta=theta)
diff = np.abs(u - precomputed[theta]).max()
# Precomputed numbers are known to 8 decimal places
nt.assert_almost_equal(diff, 0, places=8,
msg='theta=%s' % theta)
theta = 1; a = 1; I = 1; dt = 2
may lead to integer division:
(1 - (1-theta)*a*dt) # becomes 1
(1 + theta*dt*a) # becomes 2
(1 - (1-theta)*a*dt)/(1 + theta*dt*a) # becomes 0 (!)
Test that solver does not suffer from such integer division:
def test_potential_integer_division():
"""Choose variables that can trigger integer division."""
theta = 1; a = 1; I = 1; dt = 2
N = 4
u, t = decay_mod.solver(I=I, a=a, T=N*dt, dt=dt, theta=theta)
u_de = np.array([exact_discrete_solution(n, I, a, theta, dt)
for n in range(N+1)])
diff = np.abs(u_de - u).max()
nt.assert_almost_equal(diff, 0, delta=1E-14)
Convergence rate tests are very common for differential equation solvers.
def test_convergence_rates():
"""Compare empirical convergence rates to exact ones."""
# Set command-line arguments directly in sys.argv
import sys
sys.argv[1:] = '--I 0.8 --a 2.1 --T 5 '\
'--dt 0.4 0.2 0.1 0.05 0.025'.split()
r = decay_mod.main()
for theta in r:
nt.assert_true(r[theta]) # check for non-empty list
expected_rates = {0: 1, 1: 1, 0.5: 2}
for theta in r:
r_final = r[theta][-1]
# Compare to 1 decimal place
nt.assert_almost_equal(expected_rates[theta], r_final,
places=1, msg='theta=%s' % theta)
Complete program: test_decay_nose.py.
unittest is a Python module mimicing the classical JUnit
class-based unit testing framework from JavaYou will probably not use it, but you're not educated unless you know what unit testing with classes is.
Write file test_mymod.py:
import unittest
import mymod
class TestMyCode(unittest.TestCase):
def test_double(self):
result = mymod.double(4)
self.assertEqual(result, 8)
if __name__ == '__main__':
unittest.main()
import unittest
import decay_mod_unittest as decay
import numpy as np
def exact_discrete_solution(n, I, a, theta, dt):
factor = (1 - (1-theta)*a*dt)/(1 + theta*dt*a)
return I*factor**n
class TestDecay(unittest.TestCase):
def test_exact_discrete_solution(self):
...
diff = np.abs(u_de - u).max()
self.assertAlmostEqual(diff, 0, delta=1E-14)
def test_solver(self):
...
for theta in 0, 0.5, 1:
...
self.assertAlmostEqual(diff, 0, places=8,
msg='theta=%s' % theta)
def test_potential_integer_division():
...
self.assertAlmostEqual(diff, 0, delta=1E-14)
def test_convergence_rates(self):
...
for theta in r:
...
self.assertAlmostEqual(...)
if __name__ == '__main__':
unittest.main()
Complete program: test_decay_unittest.py.
Tasks:
Problem stores the physical parameters \( a \), \( I \), \( T \)
from numpy import exp
class Problem:
def __init__(self, I=1, a=1, T=10):
self.T, self.I, self.a = I, float(a), T
def u_exact(self, t):
I, a = self.I, self.a # extract local variables
return I*exp(-a*t)
Basic usage:
problem = Problem(T=5)
problem.T = 8
problem.dt = 1.5
More flexible input from the command line:
class Problem:
def __init__(self, I=1, a=1, T=10):
self.T, self.I, self.a = I, float(a), T
def define_command_line_options(self, parser=None):
if parser is None:
import argparse
parser = argparse.ArgumentParser()
parser.add_argument(
'--I', '--initial_condition', type=float,
default=self.I, help='initial condition, u(0)',
metavar='I')
parser.add_argument(
'--a', type=float, default=self.a,
help='coefficient in ODE', metavar='a')
parser.add_argument(
'--T', '--stop_time', type=float, default=self.T,
help='end time of simulation', metavar='T')
return parser
def init_from_command_line(self, args):
self.I, self.a, self.T = args.I, args.a, args.T
def exact_solution(self, t):
I, a = self.I, self.a
return I*exp(-a*t)
ArgumentParser, or make oneNone is used to indicate a non-initialized variable
class Solver:
def __init__(self, problem, dt=0.1, theta=0.5):
self.problem = problem
self.dt, self.theta = float(dt), theta
def define_command_line_options(self, parser):
parser.add_argument(
'--dt', '--time_step_value', type=float,
default=0.5, help='time step value', metavar='dt')
parser.add_argument(
'--theta', type=float, default=0.5,
help='time discretization parameter', metavar='dt')
return parser
def init_from_command_line(self, args):
self.dt, self.theta = args.dt, args.theta
def solve(self):
from decay_mod import solver
self.u, self.t = solver(
self.problem.I, self.problem.a, self.problem.T,
self.dt, self.theta)
Note: reuse of the numerical algorithm from the decay_mod module
(i.e., the class is a wrapper of the procedural implementation).
class Visualizer:
def __init__(self, problem, solver):
self.problem, self.solver = problem, solver
def plot(self, include_exact=True, plt=None):
"""
Add solver.u curve to the plotting object plt,
and include the exact solution if include_exact is True.
This plot function can be called several times (if
the solver object has computed new solutions).
"""
if plt is None:
import scitools.std as plt # can use matplotlib as well
plt.plot(self.solver.t, self.solver.u, '--o')
plt.hold('on')
theta2name = {0: 'FE', 1: 'BE', 0.5: 'CN'}
name = theta2name.get(self.solver.theta, '')
legends = ['numerical %s' % name]
if include_exact:
t_e = linspace(0, self.problem.T, 1001)
u_e = self.problem.exact_solution(t_e)
plt.plot(t_e, u_e, 'b-')
legends.append('exact')
plt.legend(legends)
plt.xlabel('t')
plt.ylabel('u')
plt.title('theta=%g, dt=%g' %
(self.solver.theta, self.solver.dt))
plt.savefig('%s_%g.png' % (name, self.solver.dt))
return plt
Remark: The plt object in plot adds a new curve to a plot,
which enables comparing different solutions from different
runs of Solver.solve
Let Problem, Solver, and Visualizer play together:
def main():
problem = Problem()
solver = Solver(problem)
viz = Visualizer(problem, solver)
# Read input from the command line
parser = problem.define_command_line_options()
parser = solver. define_command_line_options(parser)
args = parser.parse_args()
problem.init_from_command_line(args)
solver. init_from_command_line(args)
# Solve and plot
solver.solve()
import matplotlib.pyplot as plt
#import scitools.std as plt
plt = viz.plot(plt=plt)
E = solver.error()
if E is not None:
print 'Error: %.4E' % E
plt.show()
Complete program: decay_class.py.
Problem and Solver classes soon contain
much repetitive code when the number of parameters increasesself.prms,
with two associated dictionaries self.types and
self.help for holding associated object types and help stringsParametersProblem, Solver, and maybe Visualizer be subclasses
of class Parameters, basically defining self.prms, self.types,
self.help
class Parameters:
def set(self, **parameters):
for name in parameters:
self.prms[name] = parameters[name]
def get(self, name):
return self.prms[name]
def define_command_line_options(self, parser=None):
if parser is None:
import argparse
parser = argparse.ArgumentParser()
for name in self.prms:
tp = self.types[name] if name in self.types else str
help = self.help[name] if name in self.help else None
parser.add_argument(
'--' + name, default=self.get(name), metavar=name,
type=tp, help=help)
return parser
def init_from_command_line(self, args):
for name in self.prms:
self.prms[name] = getattr(args, name)
Slightly more advanced version in class_decay_verf1.py.
class Problem(Parameters):
"""
Physical parameters for the problem u'=-a*u, u(0)=I,
with t in [0,T].
"""
def __init__(self):
self.prms = dict(I=1, a=1, T=10)
self.types = dict(I=float, a=float, T=float)
self.help = dict(I='initial condition, u(0)',
a='coefficient in ODE',
T='end time of simulation')
def exact_solution(self, t):
I, a = self.get('I'), self.get('a')
return I*np.exp(-a*t)
class Solver(Parameters):
def __init__(self, problem):
self.problem = problem
self.prms = dict(dt=0.5, theta=0.5)
self.types = dict(dt=float, theta=float)
self.help = dict(dt='time step value',
theta='time discretization parameter')
def solve(self):
from decay_mod import solver
self.u, self.t = solver(
self.problem.get('I'),
self.problem.get('a'),
self.problem.get('T'),
self.get('dt'),
self.get('theta'))
def error(self):
try:
u_e = self.problem.exact_solution(self.t)
e = u_e - self.u
E = np.sqrt(self.get('dt')*np.sum(e**2))
except AttributeError:
E = None
return E
ParametersVisualizerProblem, Solver, and
VisualizerGoal: explore the behavior of a numerical method for a differential equation and show how scientific experiments can be set up and reported.
Tasks:
os.system for running other programssubprocess for running other programs and extracting the outputProblem: $$ \begin{equation} u'(t) = -au(t),\quad u(0)=I,\ 0 < t \leq T, \label{decay:experiments:model} \end{equation} $$
Solution method (\( \theta \)-rule): $$ u^{n+1} = \frac{1 - (1-\theta) a\Delta t}{1 + \theta a\Delta t}u^n, \quad u^0=I\tp $$
python decay_mod.py --I 1 --a 2 --makeplot --T 5 --dt 0.5 0.25 0.1 0.05FE_*.png, BE_*.png, and CN_*.png
to new figures with multiple plotspython decay_exper0.py 0.5 0.25 0.1 0.05 (\( \Delta t \) values on the command line)
Typical script (small administering program) for running the experiments:
import os, sys
def run_experiments(I=1, a=2, T=5):
# The command line must contain dt values
if len(sys.argv) > 1:
dt_values = [float(arg) for arg in sys.argv[1:]]
else:
print 'Usage: %s dt1 dt2 dt3 ...' % sys.argv[0]
sys.exit(1) # abort
# Run module file as a stand-alone application
cmd = 'python decay_mod.py --I %g --a %g --makeplot --T %g' % \
(I, a, T)
dt_values_str = ' '.join([str(v) for v in dt_values])
cmd += ' --dt %s' % dt_values_str
print cmd
failure = os.system(cmd)
if failure:
print 'Command failed:', cmd; sys.exit(1)
# Combine images into rows with 2 plots in each row
image_commands = []
for method in 'BE', 'CN', 'FE':
pdf_files = ' '.join(['%s_%g.pdf' % (method, dt)
for dt in dt_values])
png_files = ' '.join(['%s_%g.png' % (method, dt)
for dt in dt_values])
image_commands.append(
'montage -background white -geometry 100%' +
' -tile 2x %s %s.png' % (png_files, method))
image_commands.append(
'convert -trim %s.png %s.png' % (method, method))
image_commands.append(
'convert %s.png -transparent white %s.png' %
(method, method))
image_commands.append(
'pdftk %s output tmp.pdf' % pdf_files)
num_rows = int(round(len(dt_values)/2.0))
image_commands.append(
'pdfnup --nup 2x%d tmp.pdf' % num_rows)
image_commands.append(
'pdfcrop tmp-nup.pdf %s.pdf' % method)
for cmd in image_commands:
print cmd
failure = os.system(cmd)
if failure:
print 'Command failed:', cmd; sys.exit(1)
# Remove the files generated above and by decay_mod.py
from glob import glob
filenames = glob('*_*.png') + glob('*_*.pdf') + \
glob('*_*.eps') + glob('tmp*.pdf')
for filename in filenames:
os.remove(filename)
if __name__ == '__main__':
run_experiments()
Complete program: experiments/decay_exper0.py.
Many useful constructs in the previous script:
[float(arg) for arg in sys.argv[1:]] builds a list of real numbers
from all the command-line argumentsfailure = os.system(cmd) runs an operating system command
(e.g., another program)sys.exit(1) aborts the program['%s_%s.png' % (method, dt) for dt in dt_values] builds a list of
filenames from a list of numbers (dt_values)montage commands for creating composite figures are stored in a
list and thereafter executed in a loopglob.glob('*_*.png') returns a list of the names of all files in the
current folder where the filename matches the Unix wildcard notation
*_*.png (meaning "any text, underscore, any text, and then `.png`")os.remove(filename) removes the file with name filename
In decay_exper0.py we run a program (os.system) and
want to grab the output, e.g.,
Terminal> python decay_plot_mpl.py
0.0 0.40: 2.105E-01
0.0 0.04: 1.449E-02
0.5 0.40: 3.362E-02
0.5 0.04: 1.887E-04
1.0 0.40: 1.030E-01
1.0 0.04: 1.382E-02
Tasks:
decay_mod.py program
Use the subprocess module to grab output:
from subprocess import Popen, PIPE, STDOUT
p = Popen(cmd, shell=True, stdout=PIPE, stderr=STDOUT)
output, dummy = p.communicate()
failure = p.returncode
if failure:
print 'Command failed:', cmd; sys.exit(1)
output string, line by lineerrors with keys dt and
the three \( \theta \) values
errors = {'dt': dt_values, 1: [], 0: [], 0.5: []}
for line in output.splitlines():
words = line.split()
if words[0] in ('0.0', '0.5', '1.0'): # line with E?
# typical line: 0.0 1.25: 7.463E+00
theta = float(words[0])
E = float(words[2])
errors[theta].append(E)
Next: plot \( E \) versus \( \Delta t \) for \( \theta=0,0.5,1 \)
Complete program: experiments/decay_exper1.py. Fine recipe for